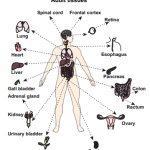
The human proteome map provides a catalog of proteins expressed in nondiseased issues and organs to use as baseline in understanding changes that occur in disease
Given the growing importance of proteins in medical laboratory testing, pathologists will want to know about a major milestone recently achieved in this field. Researchers have announced that drafts of the complete human proteome have been released to the public.
Experts are comparing this to the first complete map of the human genome that was made public in 2000. Clinical laboratory managers and pathologists know how the availability of this information provided the foundation for rapid advances in understanding different aspects involving DNA and RNA.
Map of Human Proteome Expected to Advance Medical Science
This new information about the human proteome is expected to trigger similar rapid advances in medical science and a better understanding of the underlying causes of human diseases. Two international teams recently produced the first drafts of the human protoeome, a catalog of proteins expressed in most nondiseased human issues and organs. This proteome map can be used as a baseline to understand changes that occur in the disease state, noted a report.
These studies are part of the Human Proteome Project, an international effort by the Human Proteome Organization to revolutionize our understanding of the human proteome by coordinating research at laboratories around the world directed at mapping the entire human proteome.
One Study Team Was at Johns Hopkins University
Some funding was provided by the National Institutes of Health (NIH). In one study, which was headed by Ahilesh Pandey, M.D., a researcher at Johns Hopkins University in Baltimore, and colleague Harsha Gowda, Ph.D., of the Institute of Bioinformatics in Bangalore, India, the research team used an advanced form of mass spectrometry to analyze proteins to create the human proteome map, according to a report published in NIH Research Matters.

One of two research teams, which recently published the first map of the human proteome was led by Ahilesh Pandey, M.D. (pictured above), a Professor in the Departments of Biological Chemistry, Pathology and Oncology at the Johns Hopkins School of Medicine. One primary tool used in this research was mass spectrometry. Pathologists will benefit from this investigation as the information developed by the research teams leads to the development of medical laboratory tests that improve diagnostic accuracy over existing methodologies. (Photo copyright Johns Hopkins University.)
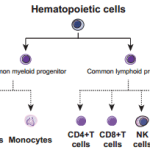
The research team examined 30 normal human tissue and cell types: 17 adult tissues, 7 fetal tissue and 6 blood cell types. Samples from three people per tissue type were processed through several steps. Next, the protein fragments, or peptides, were analyzed on high-resolution Fourier-transform mass spectrometers.The amino acid sequences were then compared to known sequences. The results were published in the May 28, 2014, issue of Nature.
The resulting draft map of the human proteome map includes proteins encoded by more than 17,000 genes, noted the Research Matters article. Among these are hundreds of proteins from regions previously thought to be non-coding.
This study also provided a new understanding of how genes are expressed. For example, almost 200 genes begin in locations other than those predicted based on genetic sequence.
“The fact that 193 of the proteins came from DNA sequences predicted to be non-coding means that we don’t fully understand how cells read DNA, because clearly those sequences do code for proteins,” said Pandey. This study also produced the Human Proteome Map, an interactive online portal. This can be accessed at this link. The study data will also soon be accessible through the National Center for Biotechnology Information.
German’s ProteomicsDB Analyzed a Mix of Available and New Tissue Data
The other study was conducted by a team lead by proteomics researcher Bernhard Küster of the Technische Universität München in Germany. Küster and his colleagues created a searchable, public database called ProteomicsDB. This database contains 92% of the estimated 19,629 human proteins, noted The Scientist article.
Küster’s team also used mass spectrometry to analyze human tissue samples. This team’s approach differed from Johns Hopkins’ in that it compiled about 60% of the information in the ProteomicsDB database by using existing raw mass spec data from databases and colleagues’ contributions.
To fill data gaps, the Küster lab generated its own mass spec data after analyzing 60 human tissues, 13 body fluids, and 147 cancer cell lines. High-resolution public data was selected and computationally processed for strict quality control, according to Küster. The database for ProteomicsDB is public and searchable. It can be accessed at this link.
German Study Added New Insights to Transcription Process
Comparing the ratio of protein to mRNA levels for every protein globally, the Küster lab found that the translation rate is a constant feature of each mRNA transcript. “This was a surprising and a really important finding,” noted Anne-Claude Gingras, Ph.D, a proteomics researcher at the Lunenfeld-Tanenbaum Research Institute in Toronto, Canada. She was quoted in The Scientist report. Gingras was not involved in either study.
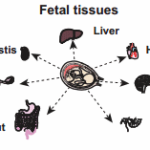
In developing the human proteome map, researcher Ahilesh Pandey, M.D., of Johns Hopkins University in Baltimore, and Harsha Gowda, Ph.D., of the Institute of Bioinformatics in Bangalore, India, analyzed tissue samples from three individuals for each tissue type in the diagrams above, including the 17 adult organs, seven fetal tissues and six blood cell types. (Images in the Public Domain)
Hanno Steen, Ph.D., Director of Proteomics at Boston Children’s Hospital, who also was not involved in the work, agreed. He explained to The Scientist, “If this observation holds true, it’s a paradigm shift. The proteomics community has viewed transcriptome and proteome data as two sides of a coin. But this analysis shows that at least, at steady state, once the ratio for an mRNA/protein pair has been calculated, protein levels can be determined just from specific mRNA levels.”
Deeper Knowledge of Proteome to Improve Diagnostics and Therapeutics
“[T]he real breakthrough with these two projects is the comprehensive coverage of more than 80% of the expected human proteome, which has not been achieved previously,” Steen concluded. “These efforts clearly show that to get to this deep level of proteome coverage, many different tissue types must be probed.”
Gingras further observed that these two studies are very complimentary. “The Hopkins group really addressed what was missing in proteomics, providing a survey of human proteins from a single source, which allows for easy comparisons within their data. In contrast, the ProteomeDB effort connected new information with existing data from the proteomics community.” Küster’s goal is to continue to grow and refine the database, further engaging the community and pooling more resources.
A deeper knowledge of the human proteome could help fill the gap between genomes and phenotypes. As this occurs, it has the potential to transform the way diagnostics and therapeutics are developed, thereby enhancing overall biomedical research and healthcare, noted a report presented to scientific leaders at a NIH workshop on advances in proteomics and its applications.
The importance of these latest studies to pathologists and Ph.D.s working in molecular diagnostics laboratories is that this information will expedite further research into the human proteome. Such research is expected to lead to novel methods of diagnosis and complex “multi-analyte” clinical laboratory tests that look for multiple proteins in a single assay.
—Patricia Kirk
Related Information: