The AI protein-structure-prediction system may ‘revolutionize life sciences by enabling researchers to better understand disease,’ researchers say
Genomics leaders watched with enthusiasm as artificial intelligence (AI) accelerated discoveries that led to new clinical laboratory diagnostic tests and advanced the evolution of personalized medicine. Now Google’s London-based DeepMind has taken that a quantum step further by demonstrating its AI can predict the shape of proteins to within the width of one atom and model three-dimensional (3D) structures of proteins that scientist have been trying to map accurately for 50 years.
Pathologists and clinical laboratory professionals know that it is estimated that there are around 30,000 human genes. But the human proteome has a much larger number of unique proteins. The total number is still uncertain because scientists continue to identify new human proteins. For this reason, more knowledge of the human protein is expected to trigger an expanding number of new assays that can be used by medical laboratories for diagnostic, therapeutic, and patient-monitoring purposes.
DeepMind’s AI tool is called AlphaFold and the protein-structure-prediction system will enable scientists to quickly move from knowing a protein’s DNA sequence to determining its 3D shape without time-consuming experimentation. It “is expected to accelerate research into a host of illnesses, including COVID-19,” BBC News reported.
This protein-folding breakthrough not only answers one of biology’s biggest mysteries, but also has the potential to revolutionize life sciences by enabling researchers to better understand disease processes and design personalized therapies that target specific proteins.
“It’s a game changer,” Andrei Lupas, PhD, Director at the Max Planck Institute for Developmental Biology in Tübingen, Germany, told the journal Nature. “This will change medicine. It will change research. It will change bioengineering. It will change everything.”
AlphaFold Wins Prestigious CASP14 Competition
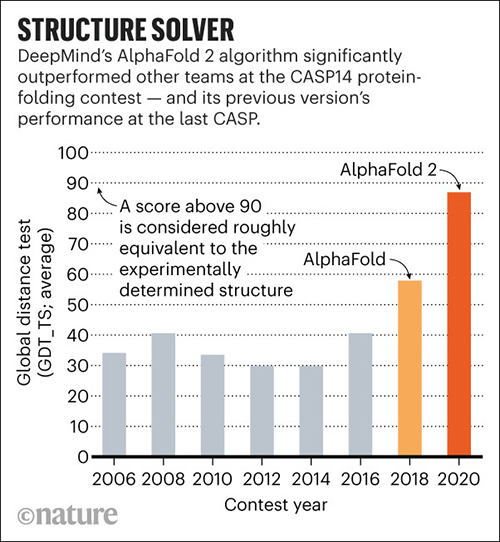
In November, DeepMind’s AlphaFold won the 14th Community Wide Experiment on Critical Assessment of Techniques for Protein Structure Prediction (CASP14), a biennial competition in which entrants receive amino acid sequences for about 100 proteins whose 3D structures are unknown. By comparing the computational predictions with the lab results, each CASP14 competitor received a global distance test (GDT) score. Scores above 90 out of 100 are considered equal to experimental methods. AlphaFold produced models for about two-thirds of the CASP14 target proteins with GDT scores above 90, a CASP14 press release states.
According to MIT Technology Review, DeepMind’s discovery is significant. That’s because its speed at predicting the structure of proteins is unprecedented and it matched the accuracy of several techniques used in clinical laboratories, including:
Unlike the laboratory techniques, which, MIT noted, are “expensive and slow” and “can take hundreds of thousands of dollars and years of trial and error for each protein,” AlphaFold can predict a protein’s shape in a few days.
“AlphaFold is a once in a generation advance, predicting protein structures with incredible speed and precision,” Arthur D. Levinson, PhD, Founder and CEO of Calico Life Sciences, said in a DeepMind blogpost. “This leap forward demonstrates how computational methods are poised to transform research in biology and hold much promise for accelerating the drug discovery process.”
Revolutionizing Life Sciences
John Moult, PhD, Professor, University of Maryland Department of Cell Biology and Molecular Genetics, who cofounded CASP in 1994 and chairs the panel, pointed out that scientists have been attempting to solve the riddle of protein folding since Christian Anfinsen, PhD, was awarded the 1972 Nobel Prize in Chemistry for showing it should be possible to determine the shape of proteins based on their amino acid sequence.
“Even tiny rearrangements of these vital molecules can have catastrophic effects on our health, so one of the most efficient ways to understand disease and find new treatments is to study the proteins involved,” Moult said in the CASP14 press release. “There are tens of thousands of human proteins and many billions in other species, including bacteria and viruses, but working out the shape of just one requires expensive equipment and can take years.”
Science reported that the 3D structures of only 170,000 proteins have been solved, leaving roughly 200 million proteins that have yet to be modeled. Therefore, AlphaFold will help researchers in the fields of genomics, microbiomics, proteomics, and other omics understand the structure of protein complexes.
“Being able to investigate the shape of proteins quickly and accurately has the potential to revolutionize life sciences,” Andriy Kryshtafovych, PhD, Project Scientist at University of California, Davis, Genome Center, said in the press release. “Now that the problem has been largely solved for single proteins, the way is open for development of new methods for determining the shape of protein complexes—collections of proteins that work together to form much of the machinery of life, and for other applications.”
Clinical laboratories play a major role in the study of human biology. This breakthrough in genomics research and new insights into proteomics may provide opportunities for medical labs to develop new diagnostic tools and assays that better identify proteins of interest for diagnostic and therapeutic purposes.
—Andrea Downing Peck
Related Information:
AI Solution to a 50-Year-Old Science Challenge Could ‘Revolutionize’ Medical Research
‘It Will Change Everything’: DeepMind’s AI Makes Gigantic Leap in Solving Protein Structures
AlphaFold: A Solution to a 50-Year-Old Grand Challenge in Biology
DeepMind’s Protein-Folding AI Has Solved A 50-Year-Old Grand Challenge of Biology
‘The Game Has Changed.’ AI Triumphs at Solving Protein Structures



