Researchers used CRISPR-based assays to develop new clinical laboratory point-of-care blood test which boasts accuracy, affordability, and accessibility
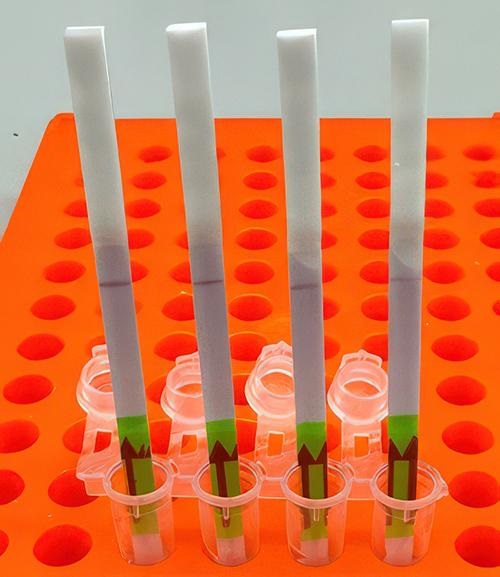
Here’s a novel use of paper as clinical laboratory test media. Researchers at Princeton University in New Jersey, the Massachusetts Institute of Technology’s Broad Institute, and Harvard University have developed an at-home paper-strip test that can not only identify the presence of influenza, but it can also differentiate between different strains of the flu bug.
According to UPI, the test can “distinguish between influenza A and influenza B—the two main types of seasonal flu—as well as identifying more virulent strains like H1N1 and H3N2.”
Many research teams are working to develop paper-based diagnostic screening tests because of their lower cost to produce and usefulness in remote locations. Should this near-patient point-of-care test become clinically viable, it could mean shorter times to answer, enabling speedier diagnoses and earlier start of treatment.
It also means patient specimens do not have to be transported to a clinical laboratory for testing. And reduced cost per test makes it possible to test more people. This serves the public health aspect of monitoring outbreaks of influenza and other diseases and gives hope for improved treatment outcomes.
“Being able to tease apart what strain or subtype of influenza is infecting a patient has repercussions both for treating them and public health interventions, said Jon Arizti Sanz, PhD, co-lead study author and postdoctoral researcher at the Broad Institute of Harvard and MIT, in a Broad Institute news release.
The researchers published their findings in The Journal of Molecular Diagnostics titled, “CRISPR-Based Assays for Point-of-Need Detection and Subtyping of Influenza.”

“Ultimately, we hope these tests will be as simple as rapid antigen tests, and they’ll still have the specificity and performance of a nucleic acid test that would normally be done in a laboratory setting,” Cameron A. Myhrvold, PhD (above), Assistant Professor of Molecular Biology at Princeton University in New Jersey, told CIDRAP. Influenza tests that can be performed at the point of care and in remote locations may reduce the number of screening tests performed by clinical laboratories. (Photo copyright: Michael James Butts/Hertz Foundation.)
Inspiration from Prior COVID-19 Test
According to an article published by the Center for Infectious Disease Research and Policy Research and Innovation Office (CIDRAP) at the University of Minnesota, the original test was developed in 2020 in a Harvard laboratory led by computational geneticist Pardis Christine Sabeti, MD, PhD, professor, Department of Organismic and Evolutionary Biology, and co-senior author of the study.
Her team developed their tests using Streamlined Highlighting of Infections to Navigate Epidemics (SHINE), “a clustered regularly interspaced short palindromic repeats (CRISPR)-based RNA detection platform,” the researchers wrote in their Journal of Molecular Diagnostics paper.
“SHINE has a runtime of 90 minutes, can be used at room temperature and only requires an inexpensive heat block to heat the reaction. The SHINE technology has previously been used to identify SARS-CoV-2 and later to distinguish between the Delta and Omicron variants,” Bioanalysis Zone reported.
“The test uses genetically engineered enzymes to identify specific sequences of viral RNA in samples,” the researchers told UPI. Originally designed to detect COVID-19, the team adapted the technology to detect influenza in 2022 “with the aim of creating a screening tool that could be used in the field or in clinics rather than hospitals or high-tech diagnostic labs,” they said.
Influenza A and B as well as H1N1 and H3N2 subtypes were the targets of the four SHINE assays. “When tested on clinical samples, these optimized assays achieved 100% concordance with quantitative RT-PCR. Duplex Cas12a/Cas13a SHINE assays were also developed to detect two targets simultaneously,” the researchers wrote in their paper.
The team used “20 nasal swabs from people with flu-like symptoms during the 2020-2021 flu season, nasal fluid from healthy people as the control, and 2016-2021 influenza sequences downloaded from the National Center for Biotechnology Information Influenza (NICB) database. They compared the results with those from quantitative reverse transcription-polymerase chain reaction (RT-PCR) tests,” CIDRAP reported.
Implications of the New Tests
The ease of the new tests is an important development since approximately only 1% of individuals who come down with the flu see doctors for testing, according to the news release. And researchers had this in mind, looking at speed, accuracy, and affordability as a means to “improve outbreak response and infection care around the world,” UPI reported.
There are great benefits to strain differentiation that be achieved with the new test. Doctors are hopeful the test will help dial in the best treatment plans for patients since some strains are resistant to the antiviral medication oseltamivir (Tamiflu), UPI noted. This is significant since Tamiflu “is a common antiviral,” said Sanz in the Broad Institute news release.
“These assays have the potential to expand influenza detection outside of clinical laboratories for enhanced influenza diagnosis and surveillance,” the Journal of Molecular Diagnostics paper noted. This allows for more strategic treatment planning.
“Using a paper strip readout instead of expensive fluorescence machinery is a big advancement, not only in terms of clinical care but also for epidemiological surveillance purposes,” said Ben Zhang, an MD candidate in the Health Sciences and Technology at Harvard and co-first author of the study, in the Broad Institute news release.
Future Plans for Tests
“With further development, the test strip could be reprogrammed to distinguish between SARS-CoV-2 and flu and recognize swine flu and avian flu, including the H5N1 subtype currently causing an outbreak in US dairy cattle,” the study authors told CIDRAP.
The team is also looking at ways to help prevent H5N1 from crossing into human contamination, Sanz told UPI.
The new Princeton/MIT/Harvard tests echo the trend to bring in affordability and ease-of-use with accurate results as an end goal. Faster results mean the best treatments for each person can start sooner and may render the transport of specimens to a clinical laboratory as a second step unnecessary.
As research teams work to develop paper-based viral tests for their plethora of benefits, clinical laboratories will want to pay close attention to this development as it can have a big implication on assisting with future outbreaks.
Additional research is needed before these tests are going to be commonplace in homes worldwide, but this first step brings inspiration and hope of what’s to come.
—Kristin Althea O’Connor
Related Information:
Simple Test for Flu Could Improve Diagnosis and Surveillance
Simple Paper-Strip Test Might Spot Flu, Identify Strain
CRISPR-Based Assays for Point-of-Need Detection and Subtyping of Influenza
Paper Strip Test Can Identify Flu Subtypes, May Have Other Applications, Scientists Say
Streamlined Inactivation, Amplification, and Cas13-based Detection of SARS-Cov-2
Paper Strip Test Using CRISPR and SHINE Technology Has Been Developed for Rapid Influenza Diagnosis



