DNA ‘dark matter’ will provide clinical laboratories and pathologists with a new specimen source and new methodology for developing useful diagnostic tests
Growing knowledge about DNA “dark matter” may soon make it possible for clinical laboratories to develop new assays that reveal clinical information useful in diagnosing and guiding therapeutic decisions. In genetics, “dark matter” describes the non-coding areas of DNA. Recent discoveries indicate that dark matter plays more important roles than previously thought.
Scientists at Vienna’s Research Institute of Molecular Pathology (IMP) recently developed a method that reliably detects the non-coding areas of DNA and simultaneously measures their activity quantitatively. The researchers then used this new methodology to study the genome of the Drosophila fly. Drosophila is widely used in genetic studies. These studies broadened the researchers’ understanding of processes such as transcription and replication in other eukaryotes, including humans. The IMP findings were published January 17, 2013, online by the journal Science.
The new insights from this study are significant for pathologists and clinical laboratory managers, particularly in the investigation of cancers, because:
- One, the ability to evaluate dark matter creates a new methodology that can be used to develop diagnostic tests;
- Two, evaluation of dark matter DNA provides a new source of diagnostic information: and,
- Third, dark matter represents a new type of specimen to examine.
‘Magic Microscope’ Sheds New Light on DNA Dark Matter
The science community once thought dark matter DNA—roughly 98% of the genome— was mere refuse from the evolutionary process. For more than a decade, however, they have recognized that these non-coding areas of DNA house gene enhancers and regulatory functions. These important regions determine where, how, and when genes are expressed, IMP stated in a press release.
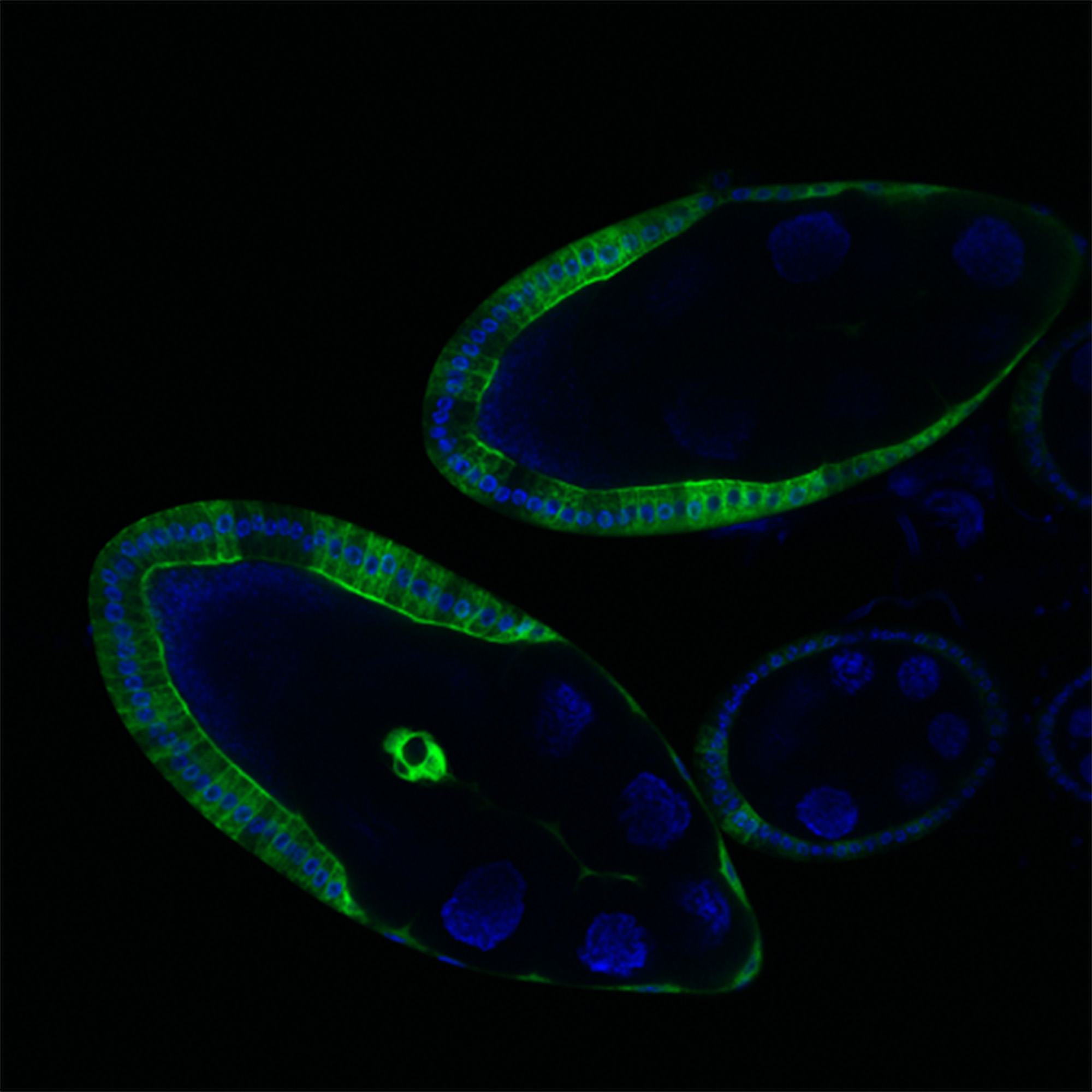
Research conducted at the Research Institute of Molecular Pathology in Vienna, Austria, has increased the knowledge of the role and functions of “dark matter” sections of DNA. Pictured above is a fluorescence image of ovarian tissue of the fruit fly. DNA is stained in blue, the activity of enhancers is represented by the green color. The extensive sections of green indicate that “dark matter” DNA is much more active than previously thought. (Photo copyright Institute of Molecular Pathology.)
Despite scientists’ better understanding of the functional importance of dark matter, the ability to study these regions remained limited. This was due to the lack of a reliable detection method, the press release stated. Researchers were constrained by the available methods of investigation. These methods were indirect and tended to be error-prone, according to IMP. Further, they required tedious experiments to validate and quantify enhancer activities.
The new technology used by the IMP researchers—dubbed STARR-seq (self-transcribing active regulatory region sequencing)—bridges the deficiency gap. It allows the direct identification of dark matter sequences and simultaneously measures their activity quantitatively in entire genomes, according to the lab website of group leader Alexander Stark, Ph.D.. “STARR-seq is like a magic microscope that lets us zoom in on the regulatory regions of DNA,” wrote the study authors in Science.
New Technology Enables Quantitative DNA Enhancer Activity Maps
The IMP scientists described the new technology as a massively parallel reporter assay based on next-generation sequencing, according to the Stark Lab website. The new method combines advanced sequencing technology and highly specialized bio-computing capabilities. “STARR-seq allows the identification of transcriptional enhancers in a direct and quantitative manner in entire genomes,” the Stark Lab website noted.
In their research, the IMP team used the new method to study the genome of the Drosophila fly. “When applied to the Drosophila genome, STARR-seq identifies thousands of cell type-specific enhancers across a broad continuum of strengths, links differential gene expression to differences in enhancer activity, and creates a genome-wide quantitative enhancer map,” they noted in the abstract. “This map reveals the highly complex regulation of transcription, with several independent enhancers for both developmental regulators and ubiquitously expressed genes.”
Scientists can use the technology to identify and quantify enhancer activity in other eukaryotes, including humans. This is especially significant in diseases such as cancer. Such diseases are clearly linked to inherited risk factors. For many of these diseases, researchers have not been able to identify the gene mutations that lead to the disease.
Mutations within the DNA enhancer system regulate gene expression and thereby the disease risk. This makes it likely that such mutations, rather than genes, will be found to play a role in those diseases. “It will be crucial to study gene regulation and how it is encoded in the genome—both during normal development and when it goes wrong in disease.” the authors wrote in Science.
Clinical Laboratories Could See New Tests for Cancer
IMP’s new method for mapping gene enhancers and regulators is expected to give pathologists a tool they can use to identify disease-creating mechanisms with far greater accuracy. In turn, this will likely spur development of new diagnostic tests and therapies, particularly in cancer care.
IMP’s new technology represents a huge leap forward in finding effective treatments to repair the underlying genetic malfunction. By extension, it is fueling hope among scientists that it will enable them to find ways to rewire broken human systems and cure and prevent cancer. At the same time, it will create opportunities for pathologists and clinical laboratory managers to provide more precise diagnostic and therapeutic information to referring physicians.
—Karen Branz
Related Information:
Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq
IMP Scientists shed light on the “dark matter” of DNA
Dark Daily, “ENCODE Project Reveals 4 Million Critical Gene Switches in ‘Junk’ DNA of Humans”


