Aug 24, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
CDC reports more than 93-million US adults are obese, and health issues related to obesity include heart disease, stroke, type 2 diabetes, and cancers
In recent years, the role of the human microbiome in weight loss or weight gain has been studied by different research groups. There is keen interest in this subject because of the high rates of obesity, and diagnostic companies know that development of a clinical laboratory test that could assess how an individual’s microbiome affects his/her weight would be a high-demand test.
This is true of a study published this year in Mayo Clinic Proceedings. Researchers at Mayo Clinic looked at obese patients who were in an active lifestyle intervention program designed to help them lose weight. It was determined that gut microbiota can have a role in both hindering weight loss and supporting weight loss.
Gut Microbiota More Complicated than Previously Thought
The Mayo researchers determined “an increased abundance of Phascolarctobacterium was associated with [successful weight loss]. In contrast, an increased abundance of Dialister and of genes encoding gut microbial carbohydrate-active enzymes was associated with failure to [lose] body weight. A gut microbiota with increased capability for carbohydrate metabolism appears to be associated with decreased weight loss in overweight and obese patients undergoing a lifestyle intervention program.”
How do bacteria impede weight loss? Vandana Nehra, MD, Mayo Clinic Gastroenterologist and co-senior author of the study, explained in a news release.
“Gut bacteria have the capacity to break down complex food particles, which provides us with additional energy. And this is normally is good for us,” she says. “However, for some individuals trying to lose weight, this process may become a hindrance.”
Put another away: people who more effectively metabolized carbohydrates were the ones who struggled to drop the pounds, New Atlas pointed out.

Vandana Nehra, MD (left), and Purna Kashyap, MBBS (right), are Mayo Clinic Gastroenterologists and co-senior authors of the Mayo study. “While we need to replicate these findings in a bigger study, we now have an important direction to pursue in terms of potentially providing more individualized strategies for people who struggle with obesity,” Nehra noted in the news release. Thus, precision medicine therapy for obese individuals could be based on Mayo Clinic’s research. (Photo copyright: Mayo Clinic.)
Mayo Study Provides Clues to Microbiota Potential in Weight Loss
The Mayo researchers wanted to know how gut bacteria behave in people who are trying to lose weight.
They recruited 26 people, ranging in age from 18 to 65, from the Mayo Clinic Obesity Treatment Research Program. Fecal stool samples, for researchers’ analysis, were collected from participants at the start of the three-month study period and at the end. The definition of successful weight loss was at least 5% of body weight.
Researchers found the following, according Live Science:
- 2 lbs. lost, on average, among all participants;
- Nine people were successful, losing an average of 17.4 lbs.;
- 17 people did not meet the goal, losing on average just 3.3 lbs.; and,
- More gut bacterial genes that break down carbohydrates were found in stool samples of the unsuccessful weight loss group, as compared to the successful dieters.
The researchers concluded that “An increased abundance of microbial genes encoding carbohydrate-active enzyme pathways and a decreased abundance of Phascolarctobacterium in the gut microbiota of obese and overweight individuals are associated with failure to lose at least 5% weight following a 3-month comprehensive lifestyle intervention program.”
Purna Kashyap, MBBS, Mayo Clinic Gastroenterologist and co-senior author of the study, told Live Science, “The study suggests there is a need to take the microbiome into account in clinical studies (on weight loss), and it also provides an important direction to pursue in terms of providing individualized care in obesity.” The very basis of precision medicine.
Future Weight-Loss Plans Based on Patient’s Microbiota
The Mayo Clinic researchers acknowledged the small sample size and need for more studies with larger samples over a longer time period. They also noted in their paper that Dialister has been associated with oral infections, such as gingivitis, and its role in energy expenditure and metabolism is unclear.
Still, the study suggests that it may soon be possible to give people individualized weight loss plans based on their gut bacteria. Clinical laboratory professionals and pathologists will want to stay abreast of follow-up studies and replication of findings by other research teams. A future medical laboratory test to analyze patients’ microbiomes could help obese people worldwide as well as lab business volume.
—Donna Marie Pocius
Related Information:
Gut Microbial Carbohydrate Metabolism Hinders Weight Loss in Overweight Adults Undergoing Lifestyle Intervention with a Volumetric Diet
Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice
CDC: Adult Obesity Facts
Makeup of an Individual’s Gut Bacteria May Play Role in Weight Loss, Mayo Study Suggests
Struggle to Lose Weight? Your gut Bacteria May Be to Blame
Your Gut Bacteria May Make It Harder to Lose Weight
Diet Hit a Snag? Your Gut Bacteria May be Partly to Blame
Can’t Lose Weight? Your Gut Bacteria Could be to Blame, According to Study
Richness of Human Gut Microbiome Correlates with Metabolic Markers
Annual Medical Spending Attributable to Obesity: Payer- and Service-Specific Estimates
5 Ways Gut Bacteria Affect Your Health
Cornell Researchers Identify Gut Microbes That May Help Some People Remain Thin; Findings Could Result in Clinical Laboratory Tests to Analyze Microbiomes of Individuals
Clinical Laboratories Might Soon be Diagnosing Obesity and Guiding Therapies that Utilize Engineered Microbes
Aug 20, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
“On-a-chip” devices continue to advance and medical laboratories will be natural repositories for patient data as the technology continues to improve
Dark Daily has predicted that the future of clinical laboratory testing will include highly complex multi-analyte test panels. The biomarkers, however, could number in the hundreds or thousands. So, it’s interesting to see new research by a Massachusetts Institute of Technology (MIT) team currently developing a multi-biomarker organ test device for clinical purposes.
Motivated by the costly failure of animal testing efforts to develop drug safety and efficacy in humans, the MIT research engineers created a microfluidic platform technology they dubbed “physiome-on-a-chip,” or more colloquially, “body-on-a-chip.” Their goal is to identify drug reaction in different cell groups within the body (in vivo).
They acknowledged contributions of in vitro microphysiological systems (MPSs), AKA “organ-on-a-chip” (OOC) systems. They note, however, in their paper published in Scientific Reports, that more complex systems that interconnect and receive data from multiple MPSs are needed due to increasing limitations arising from drugs’ “lack of efficacy” rather than toxicity.
“Here we describe the development and implementation of multi-MPS platforms, AKA physiome-on-a-chip, supporting four-way, seven-way, and 10-way MPS interactions for several weeks,” the MIT engineers wrote.
Though MIT’s new technology needs further research and development time, as well as clinical trials, this type of chip design and its ability to scale is a positive development and progress toward Dark Daily’s prediction. Once finalized, it could be adopted in medical laboratories for many types of diagnostic testing purposes.
Researchers Motivated to Improve Drug Efficacy
According to an MIT news release, “MIT engineers have developed new technology that could be used to evaluate new drugs and detect possible side effects before the drugs are tested in humans. Using a microfluidic platform that connects engineered tissues from up to 10 organs, the researchers can accurately replicate human organ interactions for weeks at a time, allowing them to measure the effects of drugs on different parts of the body.”
The “body-on-a-chip” technology, MIT says, is aimed at determining how drugs may affect one organ while also having side effects on others.
“Some of these effects are really hard to predict from animal models because the situations that lead to them are idiosyncratic. With our chip, you can distribute a drug and then look for the effects on other tissues and measure the exposure and how it is metabolized,” said Linda Griffith, PhD, Professor of Teaching Innovation at MIT’s School of Engineering, and a senior author of the study, in the news release.
According to MIT, factors affecting the effectiveness of pharmaceuticals may include:
- Genetics;
- Environment;
- Personal lifestyles; and,
- Interactions with other drugs.
TechCrunch called the study “unprecedented,” pointing to the platform’s connection of so many tissues and the technology’s ability to keep them stable for weeks.

“An advantage of our platform is that we can scale it up or down and accommodate a lot of different configurations,” Linda Griffith, PhD, MIT Professor, MIT School of Engineering, told Science Daily. “I think the field is going to go through a transition where we start to get more information out of a three-organ or four-organ system, and it will start to become cost-competitive because the information you’re getting is so much more valuable.” (Photo copyright: MacArthur Foundation.)
How “Body-on-a-Chip” Works
“Body-on-a-chip” is about the size of a tablet computer and links 10 organ types, including: liver, lung, gut, endometrium, brain, heart, pancreas, kidney, skin, and skeletal muscle.
Using microfluidic platform technology, the researchers placed one- to two-million cells from human tissue samples into the device and then pushed fluid through the chip to resemble blood flow, the Daily Mail reported, adding that MIT’s MPS platform design features:
- Compartments made from a plastic block;
- Passages for fluid to move (as a circulatory system does) between the compartments;
- A water reservoir to limit fluid evaporation; and,
- Ability to monitor flow of molecular exchanges and drug distribution.
Essentially, using the MIT device, a drug can be introduced to one organ, processed normally, and then passed to other organs for processing and use in other ways, TechCrunch summarized.
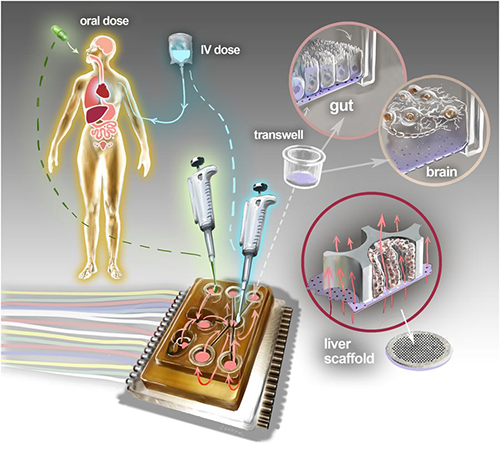
The physiome-on-a-chip system (above schematic) comprises bioengineered devices that nurture many interconnected 3D MPSs representing specified functional behaviors of each organ of interest, designed to capture essential features of in vivo physiology based on quantitative systems models tailored for individual applications such as drug fate or disease modeling. This technology could eventually be utilized for clinical laboratory and anatomic pathology testing. (Image and caption copyright: Victor O. Leshyk/Scientific Reports.)
Drug Delivery, Effects on Multiple Tissues Noted in MIT Study
The MIT researcher engineers reported these findings and accomplishments:
- Delivering a drug to the gastrointestinal tissue;
- Replicating digesting a drug;
- Observing as a drug was transported to other tissues and metabolized;
- Measuring a drug’s path; and,
- Noting effects of a drug on different tissues and how drugs break down.
“The huge potential of MPS technology is revealed by connecting multiple organ chips in an integrated system for in vitro pharmacology. This study beautifully illustrates that multi-MPS ‘physiome-on-a-chip’ approaches, which combine the genetic background of human cells with physiologically relevant tissue-to-media volumes, allow accurate prediction of drug pharmacokinetics and drug absorption, distribution, metabolism, and excretion,” said Kevin Healy, PhD, Professor of Bioengineering and Materials Science and Engineering, at University of California Berkeley in the MIT news release. Healy was not involved in the research.
Unique Device Design
In addition to making it possible to study so many different tissue types, the device design, according to MIT, is unique for these reasons:
- Its open microfluidic system, rather than a closed system, means the lid can be removed to manipulate tissue samples;
- Instead of external pumps common in closed systems, the MIT team used “on-board pumps” to control flow of liquid between the organs; and,
- The pumps used enabled larger engineered tissues, such as those from tumors in an organ, to be assessed.
The MIT engineers next plan to focus on specific organs—including the brain, liver, and gastrointestinal tissue—to model Parkinson’s disease, Digital Trends reported.
As healthcare providers and medical laboratories adopt precision medicine, MIT’s contributions are both timely and important. The ability to accommodate many different configurations in one platform is impressive, and something Dark Daily has been anticipating.
—Donna Marie Pocius
Related Information:
A “Body-on-a-Chip” Strings Together 10 Model Human Organs
“Body-on-a-Chip” Could Improve Drug Evaluation
MIT Builds “Body-on-a-Chip” Device That Can Store up to 10 Artificial Organs at Once
Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies
MIT Gadget Puts Multiple Artificial Organs into a Paperback-Sized Connected System
Drug Testing Could Get a Boost from MIT’s “Body-on-a-Chip”
Aug 10, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
This low-cost, reusable noninvasive light test could serve as a prototype for detecting other biomarkers and diseases in rural and outlying medical laboratories
A 24-year-old Ugandan computer scientist whose own malaria was missed by traditional clinical laboratory blood tests has developed a device that detects signs of the disease using a beam of light directed onto a patient’s finger. The light highlights and detects changes in the color, shape, and concentration of red bloods cells affected by disease.
Brian Gitta, CEO and co-founder of computer software company thinkIT Limited, became the youngest winner of the UK’s Royal Academy of Engineering Africa Prize for Engineering Innovation. His eHealth solution is called Matibabu, which means “treatment” in Swahili.
Gitta and his team are developing a low-cost, reusable device that clips onto a patient’s finger and provides malaria test results within 60 seconds through a mobile phone app, UPI.com reported. The latest Matibabu prototype uses a ‘hybrid of magnetic-optic technology and electro-impedance technology’ to detect the disease,” according to a blog post on the thinkIT website.
“Our next step is to determine the validity and reliability of the Matibabu device compared with the gold standard microscopy and RDT by conducting field tests with malaria patients in selected health facilities in order to obtain information that will be used to improve the device, and eventually roll it out to the market,” the blog post states.

The Matibabu malaria detection device (above) uses the principles of light scattering and magnetism to detect changes to red blood cells that signal disease. The low-cost, reusable device from thinkIT Limited has advanced through several prototypes and now has an estimated 80% accuracy rate. (Photo copyright: Makerere University College of Engineering, Design, Art and Technology.)
TechCrunch reported that the current generation of Matibabu is about 80% accurate, with the expectation that further development will increase the device’s accuracy to 90-95%.
While this new diagnostic technology needs further development and clinical studies, it could potentially be used for other biomarkers and diseases besides malaria. However, according to the Centers for Disease Control and Prevention (CDC), rapid diagnostic tests (RDTs) like this are not yet widely used, so speed-of-diagnosis also is an issue.
Nevertheless, if successful, such a non-invasive test for malaria would be a major breakthrough since, today, the mosquito-borne disease must be confirmed by medical laboratory blood tests using either microscopic diagnosis or antigen detection, which are costly and time consuming.
“It’s a perfect example of how engineering can unlock development—in this case by improving healthcare,” Rebecca Enonchong, Africa Prize judge, noted in a Royal Academy of Engineering statement. “Matibabu is simply a gamechanger.”

Africa Prize judge Rebecca Enonchong (left) presents Ugandan Brian Gitta (right) of Matibabu with the Africa Prize winner’s medal. (Photo/caption copyright: Royal Academy of Engineering.)
Shafik Sekitto, Matibabu Vice President of Business Development, told BBC World News that Gitta’s own battle with malaria was prolonged because the first three blood tests failed to diagnose his disease. “[Gitta] brought up the idea: ‘Why can’t we find a new way of using the skills we have in computer science—of diagnosing a disease without having to prick somebody?’” Sekitto explained.
Malaria Threatens Half the World’s Population
The World Health Organization (WHO) estimates that nearly half the world’s population is at risk of malaria. According to WHO estimates, in 2015 there were 429,000 deaths worldwide from malaria, with 90% of cases and 92% of deaths in sub-Saharan Africa.
The Africa Prize, which includes a $33,400 (124-million Uganda shillings) award, is Africa’s biggest prize dedicated to engineering innovation. Its sponsors aim to encourage engineers from sub-Saharan Africa “to apply their skills to develop scalable solutions to local challenges.” In addition to funding, award recipients also receive business training, mentoring, and access to the Royal Academy of Engineering’s network of high profile and experienced engineers and experts, and their networks.
Gitta expects the award of the Africa Prize will help thinkIT Limited better navigate the difficult process of gaining worldwide regulatory approval for a new diagnostic device.
“It’s such a big achievement for us, because it means that we can better manage production in order to scale clinical trials and prove ourselves to regulators,” he predicted in the Royal Academy of Engineering statement. “The recognition will help us open up partnership opportunities—which is what we need most at the moment.”
Many pathologists and clinical laboratory managers are watching the efforts of various companies to develop medical laboratory tests that can be performed with a device that is coupled to a smartphone and can be performed as a point-of-care test. A substantial proportion of these research efforts are targeting the needs for accurate diagnostic testing in developing countries. That’s because of the need for cheap, fast, and accurate assays that can be performed in the rural areas of these nations.
—Andrea Downing Peck
Related Information:
Ugandan Inventor Wins Africa Prize for Bloodless Malaria Test
Ugandan Innovation Wins the Africa Prize for Engineering Innovation
Ugandan Wins Africa Prize for Bloodless Malaria Test
Matibabu Uses Light to Diagnose Malaria
Matibabu Wins the Africa Prize for Engineering Innovation
Aug 6, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Researchers in Boston are working to develop DNA as a low-cost, effective way to store data; could lead to new molecular technology industries outside of healthcare
Even as new insights about the role of DNA in various human diseases and health conditions continue to tumble out of research labs, a potential new use for DNA is emerging. A research team in Boston is exploring how to use DNA as a low-cost, reliable way to store and retrieve data.
This has implications for the nation’s clinical laboratories and anatomic pathology groups, because they are gaining experience in sequencing DNA, then storing that data for analysis and use in clinical care settings. If a way to use DNA as a data storage methodology was to become reality, it can be expected that medical laboratories will have the skillsets, experience, and information technology infrastructure already in place to offer a DNA-based data storage service. This would be particularly true for patient data and healthcare data.
Finding a way to reduce the cost of data storage is a primary reason why scientists are looking at ways that DNA could be used as a data storage technology. These scientists and technology developers seek ways to alleviate the world’s over-crowded hard drives, cloud servers, and databases. They hope this can be done by developing technologies that store digital information in artificially-made versions of DNA molecules.
The research so far suggests DNA data storage could be used to store data more effectively than existing data storage solutions. If this proves true, DNA-based data storage technologies could play a key role in industries outside of healthcare.
If so, practical knowledge of DNA handling and storage would be critical to these companies’ success. In turn, this could present unique opportunities for medical laboratory professionals.
DNA Data Storage: Durable but Costly
Besides enormous capacity, DNA-based data storage technology offers durability and long shelf life in a compact footprint, compared to other data storage mediums.
“DNA has an information-storage density several orders of magnitude higher than any other known storage technology,” Victor Zhirnov, PhD, Chief Scientist and Director, Semiconductor Research Corporation, told Wired.
However, projected costs are quite high, due to the cost of writing the information into the DNA. However, Catalog Technologies Inc. of Boston thinks it has a solution.
Rather than producing billions of unique bits of DNA, as Microsoft did while developing its own DNA data storage solution, Catalog’s approach is to “cheaply generate large quantities of just a few different DNA molecules, none longer than 30 base pairs. Then [use] billions of enzymatic reactions to encode information into the recombination patterns of those prefab bits of DNA. Instead of mapping one bit to one base pair, bits are arranged in multidimensional matrices, and sets of molecules represent their locations in each matrix.”
The Boston-based company plans to launch an industrial-scale DNA data storage service using a machine that can daily write a terabyte of data by leveraging 500-trillion DNA molecules, according to Wired. Potential customers include the entertainment industry, federal government, and information technology developers.
Catalog is supported by $9 million from investors. However, it is not the only company working on this. Microsoft and other companies are reportedly working on DNA storage projects as well.

“It’s a new generation of information storage technology that’s got a million times the information density, compared to flash storage. You can shrink down entire data centers into shoeboxes of DNA,” Catalog’s CEO, Hyunjun Park, PhD (above center, between Chief Science Officer Devin Leake on left and Milena Lazova, scientist, on right), told the Boston Globe. (Photo copyright: Catalog.)
Microsoft, University of Washington’s Synthetic DNA Data Storage
Microsoft and researchers at the University of Washington (UW) made progress on their development of a DNA-based storage system for digital data, according to a news release. What makes their work unique, they say, is the large-scale storage of synthetic DNA (200 megabytes) along with the ability to the retrieve data as needed.
“Synthetic DNA is durable and can encode digital data with high density, making it an attractive medium for data storage. However, recovering stored data on a large-scale currently requires all the DNA in a pool to be sequenced, even if only a subset of the information needs to be extracted,” the researchers wrote in their paper published in Nature Biotechnology.
“Here, we encode and store 35 distinct files (over 200 megabytes of data ) in more than 13-million DNA oligonucleotides and show that we can recover each file individually and with no errors, using a random access approach,” the researchers explained.
“Our work reduces the effort, both in sequencing capacity and in processing, to completely recover information stored in DNA,” Sergey Yekhanin, PhD, Microsoft Senior Researcher, told Digital Trends.
Successful research by Catalog, Microsoft, and others may soon lead to the launch of marketable DNA data storage services. And medical laboratory professionals who already know the code—the life code that is—will likely find themselves more marketable as well!
—Donna Marie Pocius
Related Information:
The Rise of DNA Data Storage
The Next Big Thing in Data Storage is Actually Microscopic
Catalog Hauls in $9 Million to Make DNA-Based Data Storage Commercially Viable
UW and Microsoft Researchers Achieve Random Access in Large-Scale DNA Data Storage
Random Access in Large-Scale DNA Data Storage
Microsoft and University of Washington Show DNA Can Store Data in Practical Way
Aug 1, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
Identifying patients who will likely develop prolonged concussion symptoms could lead to new clinical laboratory tests and personalized medicine treatments
Researchers are homing in on a new diagnostic assay for concussion that could potentially generate significant numbers of test referrals to the nation’s clinical laboratories. This innovative work is targeting how concussions are diagnosed and treated.
Each year, thousands of children receive sports-related injuries, including concussions. There are ways for anatomic pathologists and hospital medical laboratories to diagnose concussions; however, testing can be invasive and doesn’t always reveal a complete picture of the injury state.
Additionally, about one third of children with concussions develop prolonged symptoms. However, when prescribing treatment plans, physicians have been unable to predict which patients are likely to recover quickly versus those who will have a longer recovery.
Now, researchers at Penn State College of Medicine (Penn State) believe they have discovered five microRNAs in saliva that could be used to identify patients who will likely experience prolonged concussion symptoms even one month after the initial injury.
The study also found that certain materials in saliva can help diagnose the severity of concussions and could hold the key to more effective clinical laboratory tests and personalized medicine treatments.
The Penn State researchers published their study results in JAMA Pediatrics, a publication of the Journal of the American Medical Association (JAMA).
Concussion Leading Sports-related Brain Injury
There are approximately 3.8 million sports and recreation-related traumatic brain injuries in the United States each year and the majority of those cases are concussions, according to The Concussion Place. Most concussions treated in emergency rooms are due to falls, motor-vehicle related injuries, being struck by an object, assaults, or playing sports.
Also known as mild traumatic brain injuries (mTBI), concussions are caused by blows or jolts to the head or body that cause the brain to move with excessive force inside the skull. The sudden impact damages brain cells and causes chemical changes within the brain that alter normal functioning. Though usually not life threatening, the damage can be serious and linger for months.
Symptoms of concussion include: headaches, fatigue, nausea, vomiting, dizziness, balance problems, confusion, memory problems, sleep disturbances, and double or blurry vision. Symptoms usually occur immediately, but could take days or even weeks to appear.
Identifying Severity/Predicting Prolonged Symptoms of Traumatic Brain Injuries
After a concussion occurs, brain cells release small fragments of genetic material known as microRNAs while they attempt to repair themselves. A portion of these microRNAs appear in the injured person’s blood and saliva.
In order to determine whether these microRNAs could be used to determine the severity of a traumatic brain injury and predict whether prolonged symptoms would occur, the prospective cohort study researchers gathered saliva samples from 52 concussion patients between the ages of seven and 21:
- The average age of the subjects was 14;
- Twenty-two of the participants were female;
- They were all athletes; and,
- The majority of the samples were collected one to two weeks after the initial injury.
The researchers examined distinct microRNAs in the samples and identified some that enabled them to predict how long a patient’s concussion symptoms might last. In addition, they found one microRNA in children and young adults that accurately predicted which subjects would experience memory and problem-solving difficulties as part of their symptomatology.
The researchers also evaluated the concussion patients using the Sport Concussion Assessment Tool (SCAT-3), Third Edition. Physicians use this questionnaire to assess the symptoms and severity of concussions. The researchers also asked the parents of the concussed patients for observations about their children’s symptoms.
During follow up visits, which occurred at four- and eight-week increments following the original assessment, the Penn State researchers collected additional saliva samples and re-evaluated the patients using SCAT-3.
New Biomarkers Based on MicroRNAs Instead of Protein
“There’s been a big push recently to find more objective markers that a concussion has occurred, instead of relying simply on patient surveys,” Steven Hicks, MD, PhD, Assistant Professor of Pediatrics, Penn State College of Medicine, Hershey, Pa., one of the study researchers, told Penn State News.
“Previous research has focused on proteins, but this approach is limited because proteins have a hard time crossing the blood-brain barrier. What’s novel about this study is we looked at microRNAs instead of proteins, and we decided to look in saliva rather than blood,” he noted.

According to Steven Hicks, MD, PhD (above), who worked on the Penn State College of Medicine study, microRNAs could be more accurate than the traditional questionnaire when diagnosing and forecasting the effects of concussions. “The microRNAs were able to predict whether symptoms would last beyond four weeks with about 85% accuracy,” he told Penn State News. “In comparison, using the SCAT-3 report of symptoms alone is about 64% accurate. If you just go off the parent’s report of symptoms, it goes down to the mid-50s. In this pilot study, these molecular signatures are outperforming survey tools.” (Photo copyright: MD Magazine.)
The goal of this research was to develop a way to definitively ascertain that a concussion had occurred, predict the length and type of symptoms, and then use that data to improve and personalize care for children and young adults who have had a concussion.
“With that knowledge physicians could make more informed decisions about how long to hold a child out of sports, whether starting more aggressive medication regimens might be warranted, or whether involving a concussion specialist might be appropriate,” Hicks told MD Magazine. “Anytime we can use accurate, objective measures to guide medical care, I think that represents an opportunity to improve concussion treatment.”
Further research and clinical trials will be needed to solidify the effectiveness and accuracy of these new biomarkers. However, a rapid, non-invasive saliva test that can determine the severity of a concussion, and predicted whether prolonged symptoms will likely occur, would be widely used and could be an important assay for clinical laboratories. Particularly those associated with hospital medical laboratories and emergency rooms.
—JP Schlingman
Related Information:
Association of Salivary MicroRNA Changes with Prolonged Concussion Symptoms
Saliva Test May Detect Biomarker for Prolonged Concussion
Molecules in Spit May be Able to Diagnose and Predict Length of Concussions
Prolonged Concussion Symptoms Identifiable by Salivary MicroRNA
Spit Test May Help Reveal Concussion Severity
Saliva Test May Lead to Improved Concussion Care for Youths
Jul 27, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Pathology, Management & Operations, News From Dark Daily
Tuft’s proof-of-concept demonstration study shows how changes in saliva can be employed as biomarkers for development of future diagnostic monitoring devices and applications
For years, pathologists and dentists have recognized that the mouth contains many useful biomarkers for a wide range of health conditions and diseases. Now a study by a research team at Tufts University School of Engineering (Tufts) has demonstrated that a tooth-mounted sensor can reliably measure certain target markers.
In this proof-of-concept study, Tufts researchers developed a tooth-mounted sensor that monitors food consumption as it enters the body. This potentially adds behavioral data to the growing list of exploitable biomarkers available to developers of in vitro diagnostics (IVDs) and wearable medical monitoring devices. For that reason, many clinical laboratory managers and anatomic pathologists will want to track further development of this technology, which uses the mouth as the source of the markers to be measured.
A report detailing the device was first published in the scientific journal Advanced Materials in March of this year.
Sensor Reacts to Biomarkers in Saliva
The 2×2-millimeter flexible sensor consists of three layers and adheres to the tooth like a sticker. It has two gold outer rings surrounding an inner layer of bio-responsive material that is highly sensitive to glucose, salt, and alcohol. The presence of any of these substances alters the electrical properties of the sensor and incites it to transmit radio frequency waves that can be received by mobile devices.
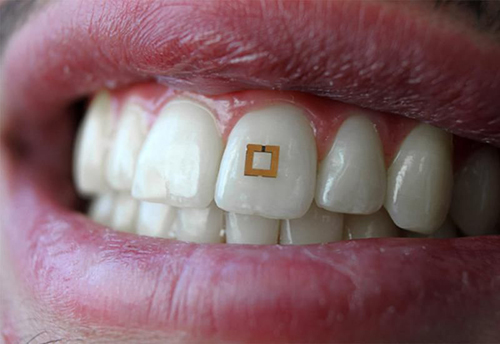
Researchers conducting a proof-of-concept study at Tufts University School of Engineering have developed “a materials‐based strategy to add utility to traditional dielectric sensors by developing a conformal radiofrequency (RF) construct composed of an active layer encapsulated between two reverse‐facing split ring resonators,” their paper published in Advanced Materials notes. The sensor is shown above mounted to a tooth, where it reacts to the presence of certain biomarkers in the saliva, triggering the transmission of an RFID signal. This device has the potential to also measure the same biomarkers used in clinical laboratory tests. (Photo copyright: Smithsonian Magazine/Tufts University School of Engineering.)
There are many possible uses for this tooth-mounted sensor. Individuals with medical conditions such as diabetes, celiac disease, or hypertension, which require them to avoid certain substances in their diet, could benefit from utilizing a device that employs the technology under development at Tufts.
Such a gadget might also help those trying to lose weight. The creators hope to enhance the material, so it has the ability to discern additional nutrients and chemicals.
“If you can evolve the sensor and engineer it to have a database of food consumption, then you could think about nutrition management,” Fiorenzo Omenetto, PhD, Professor, Department of Biomedical Engineering at Tufts and one of the authors of the research told Smithsonian Magazine. “That could be reminding us that we’re indulging too much in sugar or something like that.”
It also could potentially detect physiological or chemical changes taking place in the body by detecting certain bio-markers in the saliva.
“In theory we can modify the bio-responsive layer in these sensors to target other chemicals. We’re really limited only by our creativity,” Omenetto noted in a news release. “We have extended common RFID [radio frequency identification] technology to a sensor package that can dynamically read and transmit information on its environment, whether it is affixed to a tooth, to skin, or any other surface.”
Other Food Intake Devices
There have been previous attempts to develop wearable devices that monitors food intake. However, those gadgets usually required the use of mouth guards and head gear, which are too cumbersome for continuous everyday use. The minute size of the Tufts tooth-mounted device renders it more practical for consumers. And, since it can be mounted anywhere on a tooth—front or back—it can be made undetectable while being worn.
“This study is an interesting proof-of-concept demonstration that small, wireless biosensors can detect changes in saliva due to the presence of compounds such as salt, sugar, and alcohol,” Ben Almquist, PhD, a lecturer in the Department of Bioengineering at Imperial College London, told Smithsonian Magazine.
“For instance, for continuous monitoring of food intake, the sensors will need to be robust enough to withstand abrasion during chewing,” Almquist noted. “In addition, foods are complex mixtures of compounds including salts, sugars and proteins, and the relative amounts of each that enter into saliva will depend on factors such as the nature of the food [i.e., cooked versus fresh], the amount of chewing, and the time in the mouth before swallowing.”
The device currently remains in the prototype stage and more testing will be needed to determine its efficacy and durability. However, the emergence of such wearable devices for medical use suggests valuable opportunities for clinical laboratories.
Because data captured from the tooth-mounted device is transmitted wirelessly, clinical laboratories could potentially store and monitor the data, compare the collected data to other medical laboratory test results for the same patient, then communicate that information to clinicians, other caregivers, and even the patients. This would be a new way for clinical laboratories to provide innovative, value-added services to healthcare professionals and consumers.
—JP Schlingman
Related Information:
This Tiny Tooth Sensor Could Keep Track of the Food You Eat
Scientists Develop Tiny Tooth-mounted Sensors That Can Track What You Eat
A New Tooth-mounted Sensor Will Soon Help You Lose Weight
Functional, RF‐Trilayer Sensors for Tooth‐Mounted, Wireless Monitoring of the Oral Cavity and Food Consumption











