Feb 21, 2018 | Instruments & Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
It has been regularly demonstrated in recent decades that human breath contains elements that could be incorporated into clinical laboratory tests, so the decision to use this “breath biopsy” test in a therapeutic drug trial will be closely watched
When a major pharma company pays attention to a breath test, implications for clinical laboratories are often forthcoming. Such may be the case with GlaxoSmithKline (GSK). The global healthcare company has selected Owlstone Medical’s Breath Biopsy technology for use in its Phase II clinical trial of danirixin (DNX), a respiratory drug under development by GSK for treatment of chronic obstructive pulmonary disease (COPD), an Owlstone Medical news release announced.
Anatomic pathologists and medical laboratory leaders will be intrigued by GSK’s integration of breath-based specimens in a clinical trial of a respiratory drug. The partners in the trial aim to analyze breath samples to better understand the drug’s treatment effects and to discover personalized medicine (AKA, precision medicine) opportunities.
GSK (NYSE:GSK), headquartered in the UK but with a large presence in the US, researches and develops pharmaceutical medicines, vaccines, and other consumer health products.
Owlstone Medical, a diagnostic company, is developing a breathalyzer for disease and says it is on a mission to save 100,000 lives and $1.5 billion in healthcare costs. Dark Daily previously reported on Owlstone Medical’s Breath Biopsy platform. The Cambridge, England-based company has raised significant funding ($23.5 million) and already garnered credible cancer trial collaborators including the UK’s National Health Service (NHS).
Now, Owlstone Medical has brought its breath analysis technology to bear on chronic disease outside of cancer diagnostics development. A pharmaporum article called Owlstone’s Medical’s work with GSK an “additional boost of confidence” in the company’s technology, as well as a means for revenue.

Billy Boyle, co-founder and Chief Executive Officer, Owlstone Medical (above), shown with the company’s ReCIVA Breath Sampler device. This will be used by GSK in its Phase II respiratory disease clinical trial of danirixin to “capture VOC biomarkers in breath samples.” (Photo copyright: Business Weekly UK.)
GSK Studying Future Treatments for Respiratory Diseases
COPD affects about 700 million people worldwide, an increase of about 65% since 1990, GSK pointed out. In September 2017, GSK presented respiratory disease data and its pipeline medications at the European Respiratory Society in Milan, Italy. Included was information on danirixin (an oral CXCR2 antagonist), which is part of the company’s focus on COPD disease modification, according to a GSK news release.
“Each of our studies sets the bar for our future research and innovation,” noted Neil Barnes, MA Cantab, FRCP, FCCP(Hon), Vice President, Global Franchise Medical Head, GSK Respiratory, in the GSK press release.
Clinical Trial Aimed at Identifying the ‘Right’ Patients
With Owlstone Medical’s breathalyzer, GSK plans to explore how volatile organic compounds (VOCs) can help identify patients who will benefit most from the company’s medications, as well as evaluate Danirixin’s effects. A critical element of personalized medicine.
“It’s part of our efforts to identify the right patient for the right treatment,” said Ruth Tal-Singer, PhD, GSK’s Vice President of Medicine Development Leader and Senior Fellow, Respiratory Research and Development, in the Owlstone Medical news release.
VOCs in breath will be captured in a non-invasive way from patients who wear Owlstone Medical’s ReCIVA Breath Sampler, which, according to Owlstone Medical, has CE-mark clearance, a certification noting conformity with European health and safety standards. The VOCs breath samples will then be sent to Owlstone Medical’s lab for high-sensitivity analysis.
“Non-invasive Breath Biopsy can establish a role in precision medicine applications such as patient stratification and monitoring treatment response,” said Billy Boyle, Owlstone Medical’s co-Founder and Chief Executive Officer.
VOC Biomarkers in Respiratory Disease
VOC profiles can be characteristic of COPD as well as other respiratory diseases including asthma, tuberculosis, and cystic fibrosis, reported Science/Business.
According to Owlstone Medical’s Website, VOCs are gaseous molecules produced by the human body’s metabolism that are suitable for Breath Biopsy. Their research suggests that exhaled breath reflects molecular processes responsible for chronic inflammation. Thus, VOCs captured through Breath Biopsy offer insight into respiratory disease biomarkers.
Breath also includes VOCs that originate from circulation, which can provide information on a patient’s response to medications.
How the Breath Biopsy Platform Works
Owlstone Medical’s platform relies on its patented Field Asymmetric Ion Mobility Spectrometry (FAIMS) technology, which “has the ability to rapidly monitor a broad range of VOC biomarkers from breath, urine and other bodily fluids with high sensitivity and selectivity,” according to the company’s website. During the process:
- Gases are exchanged between circulating blood and inhaled fresh air in the lungs;
- VOC biomarkers pass from the circulation system into the lungs along with oxygen, carbon dioxide, and other gases;
- Exhaled breath contains exiting biomarkers.
It takes about a minute for blood to flow around the body. So, a breath sample during that time makes possible collection and analysis of VOC biomarkers from any part of the body touched by the circulatory system.
The medical analysis is enabled by software in the Owlstone Medical lab, Boyle told the Cambridge Independent.
“There’s an analogy with blood prints—you get the blood and can look for different diseases, and we’ve developed core hardware and technology to analyze the breath sample,” he said.
Another Breath Sample Device
The ReCIVA Breath Sampler is not the only breathalyzer focused on multiple diseases. Dark Daily reported on research conducted by Technion, Israel’s Institute of Technology, into a breath analyzer that can detect up to 17 cancers, and inflammatory and neurological diseases.
But Owlstone Medical stands out due, in part, to its noteworthy partners: the UK’s National Health Service, as well as the:
And now the company can add collaboration with GSK to its progress. Though some question the reliability of breath tests as biomarkers in the areas of sensitivity and specificity required for cancer diagnosis, Owlstone Medical appears to have the wherewithal to handle those hurdles. It is a diagnostics company that many pathologists and medical laboratory professionals may find worth watching.
—Donna Marie Pocius
Related Information:
Owlstone Medical’s Breath Biopsy Platform Integrated into GSK’s Phase II Respiratory Disease Clinical Trial
GSK Utilizes Owlstone Disease Breathalyser for Key Clinical Trials
GSK Presents Respiratory Data from Pipeline to Clinical Practice at ERS
GSK Boosts Medtech First Owlstone with Use of Breath Biopsy in Respiratory Trial
Glaxo to Stratify COPD Trial Using Breath Biopsy Device
Billy Boyle of Owlstone Medical on the Inspiration Behind His Mission to Save 100,000 From Dying of Cancer
Owlstone Medical and UK’s NHS Study Whether Breath Contains Useful Biomarkers
Breath Based Biomarker Detection: Informing Drug Development and Future Treatment Regimes
Clinical Laboratories Could Soon Diagnose 17 Diseases with a Single Breath Analyzer Test from Israel’s Institute of Technology
Feb 16, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Pathology, Management & Operations
Precision medicine programs can benefit from wearable usage data; however, little information has been collected on personalities and behaviors of the device users
Wearables medical devices have the potential to monitor some of the same biomarkers used in medical laboratory tests today. In addition, these mobile technologies can make it possible for clinical laboratories to monitor patients in real time, as well as allow labs to incorporate such into a patient’s historical record of lab test results.
The trend toward personalized medicine (aka, Precision Medicine) is increasing, with many payment programs based on it. Thus, monitoring and correcting activities that cause chronic disease, or work against treatments, is becoming standard procedure for forward-thinking, technically proficient doctors and hospitals. But are patients onboard with all of it?
Activity Trackers for Monitoring Patient Behavior
With the popularity of activity trackers on the rise, researchers are examining their usage patterns to determine how the devices are being utilized, their target market, and ways to encourage sustained use of the gadgets.
A recent article published in Annals of Internal Medicine provided insight regarding who is using this type of wearable device, how activity trackers are being employed, and the length of time consumers will maintain their usage.
The research was spearheaded by Mitesh Patel, MD, Assistant Professor of Medicine and Health Care Management, Perelman School of Medicine and the Wharton School, University of Pennsylvania. He believes this is the largest study of its kind to evaluate the usage of wearable fitness trackers.
“Many people are excited by the potential of using activity trackers to monitor healthy behaviors, but there is very little evidence on who is using them and whether or not use is sustained over time,” Patel stated in a Penn Medicine news release. “We found that, though use grew over time, it really varied depending on individual characteristics like age and income. We also found that once someone started using an activity tracker, sustained use at six months was high at 80%.”

Patel is also Director of the Penn Medicine Nudge Unit, a behavioral design team that is studying the impact that nudges or small interventions may have on healthcare. The team is examining ways in which nudges can influence choices, and also direct medical professionals and patients toward optimal decisions to improve healthcare delivery and results. (Photo copyright: University of Pennsylvania.)
Gaming the Study Improves Usage of Test Devices
To perform the study, 4.4 million members of a national wellness program were invited to take part in data collection. Approximately 55,000 of those individuals actually participated in the study, which involved downloading an app to record pertinent information. Researchers tracked and interpreted the data during a two-year period in 2014 and 2015.
The information analyzed included:
- When participants initially activated their tracker;
- How often the device was utilized;
- The average number of steps taken per day; and,
- Sociodemographic characteristics.
The results of the study were not entirely unexpected, but there were surprises:
- 80% of the people who initially activated the devices were still using them after six months;
- Only 0.2% of the invited individuals used the devices in the first year;
- However, that number increased to 1.2% during the second year.
The usage of wearable activity trackers was nearly double among younger people than it was for older individuals. In addition, people from households with an annual income of less than $50,000 used the gadgets at lower rates than those at higher income levels.
A mere 0.1% of the potential participants were over 65-years old. However, 90% of individuals in this age group were still using the devices six months after initial activation.
The authors of the study stated that adding game elements, such as points, levels, badges and financial incentives may have played a role in the sustained use of the activity trackers.
“Gamification and financial incentives are commonly used within wellness programs, but their impact has not been well studied,” Patel stated in the news release. “Our findings provide initial evidence suggesting that these types of engagement strategies may show promise for keeping sustained use high. However, more studies are needed to determine the best way to combine these types of engagement strategies with activity trackers to improve health outcomes.”
Most Commonly Used Mobile Activity Tracking Devices
There were 60 different types of wearable activity trackers that could be used by participants for the study. Seventy-six percent of those participants elected to use the FitBit activity tracker. This mobile healthcare device is worn on the wrist like a watch. It monitors activity, exercise, food, weight, and sleep to provide consumers with real-time data about their activities.
The data collected by the device is sent automatically and wirelessly to the user’s phone or computer. Individuals then can use the FitBit dashboard to view their progress through online charts and graphs. The dashboard also offers progress notifications to the consumer and gives achievement badges when established goals have been reached.
The second most common activity trackers used were Apple devices, such as Apple Watches, which were chosen by 9% of the participants.
Biometric data on patients’ behavior and activities that is collected and transmitted from mobile devices has swiftly become critical data doctors use in precision medicine diagnoses and treatments. Clinical laboratories will likely be including biomarker data taken by these devices in their testing and procedures in the future. The only question is how quickly the data generated by such devices becomes acceptable to add to a patient’s permanent health record.
—JP Schlingman
Related Information:
New Wellness Study Shows Just How Sticky Wearables Can Be, Even Among Seniors
Penn Study Shows 80% of Activity Tracker Users Stick with the Devices for at Least Six Months
Game Time: To Increase Exercise, Study Shows Gaming Strategies and a Buddy Are Key
When Push Comes to Nudge
Improvements to Fitness Wearables Help Stream Data from Consumers’ Homes to EHRs and Clinical Pathology Laboratories
Apple May Be Developing Mobile Device Technology to Monitor User’s Health and Transmit Data in Real Time
Feb 12, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Doctors’ advocacy organization praises potential of ‘My Health Record’ but voices concerns about functionality, interoperability, and added burden placed on providers
Australia’s goal of implementing a nationwide electronic health record (EHR) system received a major boost when the country’s largest pathology laboratories signed agreements with the Australian Digital Health Agency (ADHA) to join the project. But the My Health Record system has yet to fully win over providers as the Australian Medical Association (AMA) raises concerns over functionality, interoperability, and the added burden placed on healthcare providers.
ADHA Chief Executive Tim Kelsey praised the addition of pathology and diagnostic organizations to the My Health Record platform. In Australia, pathology laboratory is the term to describe what are called clinical laboratories in the United States.
“The largest diagnostic organizations in Australia have now agreed to share their test reports with Australian consumers,” Kelsey said in an ADHA news release. “We are working to deliver a My Health Record for all Australians next year, unless they choose not to have one. Health consumers will benefit from this significant commitment by the pathology industry and their software partners.”

Australian Digital Health Agency CEO Tim Kelsey says his agency in 2018 will be creating a universal electronic health record for all the country’s 24.8 million citizens, though patients will have the option to opt out of the My Health Record project. He called the “significant commitment” of pathology labs to the project a major step forward. (Photo copyright: ADHA.)
In May 2017, Sonic Healthcare, Australia’s largest pathology provider, became the first private pathology company to join the My Health Record initiative. That news was followed by agreements between the ADHA and pathology companies Primary Health Care, Australian Clinical Labs, and seven other software vendors and pathology laboratories, including:
The ADHA also finalized service agreements with additional software companies that will enable diagnostic imaging providers to link up to My Health Record by the end of 2018.
AMA Says My Health Record Lacks Functionality and Critical Features
In 2012, Australia announced the roll out of the Personally Controlled Electronic Health Record, the original initiative to create a citizen-controlled secure online summary of health information, which later was renamed My Health Record. According to The Australian, more than 5.3 million Australians are now using My Health Record, a 500% increase in the number of shared health summaries uploaded in 2016-2017 and a 200% rise in interoperability with private hospitals.
Royal College of Pathologists of Australasia President Bruce Latham, MBBS, welcomed the announcement of the increased functionality for My Health Record.
“The Australian pathology sector has been working in support of the national eHealth agenda for a number of years,” Latham stated in the ADHA news release. “Work is now progressing to connect both public and private labs to the My Health Record, and patients nationally will start to see their pathology reports in their My Health Record.”
Developers and program administrators of My Health Record predict it will generate savings of AU$123 million from:
- Reduction in adverse drug events;
- Fewer duplicated diagnostics tests; and,
- Cost savings by 2020-2021.
However, the AMA, Australia’s doctors’ advocacy group, outlined its concerns about My Health Record in a Pre-Budget Submission to the Australian federal government. While praising the project’s potential to “not only save money, but save lives,” the AMA argued the national repository of healthcare information needs improved features and functionality to meet its potential.
“… more work is required,” the AMA wrote. “The return on investment will hinge in the short term on ease of use for medical practitioners who upload the clinical data. Interoperability with the multiple software packages used across the medical profession and broader health sector must be seamless.
“Problems uploading specialists’ letters, poor search functionality, time-consuming adaptations to existing medical practitioner work practices, or inappropriate workarounds will erode clinical utility and deter doctor use—and, more importantly, take time away from focusing on the patient,” the AMA concluded.
Automatic Enrollment Concerns AMA
My Health Record began as a self-register model, but as the program goes nationwide in 2018, it will do so using an “opt-out” model. This means citizens will be enrolled automatically unless they ask to be removed from the program. According to the ADHA, the automatic creation of My Health Record for all Australians will begin in mid-2018. The government’s goal is to provide access to My Health Record to all 24.6 million Australians by June 30, 2018.
The federal government’s switch to an opt-out system for My Health Record drew concerns from the AMA.
“Doctors do not have time to talk their patients through the My Health Record arrangements for opt-out, privacy, [or] setting access controls in standing consent for health providers to upload health information. This is the work of the government. Doctors must be allowed to focus on what they do best—caring for patients,” the AMA stressed.
Clinical Laboratories Have Stake in Outcome
According to Healthcare IT News Australia, the Australian government has spent AU$2 billion ($1.53 billion USD) so far developing what could become a white elephant if general practitioners and hospital groups don’t see a clinical benefit in its use.
If Australia is successful in creating a fully-functioning and widely-used national repository for health information, it will be among the first countries to do so. In 2002, the United Kingdom (UK) kicked off a nearly decade-long effort to create a national EHR system for the UK’s single-payer tax-supported health system. Ultimately, the government pulled the plug on the initiative after spending 12.7 billion pounds ($17 billion USD) trying to complete the project.
That result, and lessons learned from Australia’s experience, should inform American healthcare policy makers. It remains a daunting effort to implement a single electronic patient health records system. Of course, pathologists and clinical laboratory administrators have an interest in this issue, since medical laboratory test data represents the largest proportion of an individual patient’s permanent health record.
—Andrea Downing Peck
Related Information:
Australia’s Largest Pathology Labs Sign Up to My Health Record
Private Pathology Reports to Go Live in My Health Record
E-health Revolution Gather Pace as more Services Pledge their Backing
Australian Medical Association Pre-Budget Submission 2018-19
National Expansion of My Health Record in 2018 Confirmed in Budget Announcement
GPS and Hospitals Claim My Health Record Not Fit for Purpose as Alarming Low Usage Figures Are Released
Feb 5, 2018 | Digital Pathology, Instruments & Equipment, Laboratory News, Laboratory Pathology, Laboratory Testing, Management & Operations
Research published in JAMA Pediatrics reports that non-invasive salivary microRNA testing identifies prolonged concussion symptoms with 85% accuracy
Sports-related concussions are always tragic, but doubly so when they involve child athletes. Quick diagnoses and treatments are critical to prevent permanent brain injury. But doctors are often hampered by the pace at which traditional medical imaging modalities and clinical laboratory diagnostic technologies provide crucial feedback.
Now, researchers at Penn State Health Children’s Hospital have determined that microRNA in saliva could be used as biomarkers in point-of-care concussion testing during sports events, according to a Penn State Health news release. Such sideline saliva analyses could provide quick feedback to field doctors on whether a head injury is serious enough to put injured athletes out of play, and how long the effects of such injuries might last. But is it accurate?
Jeremiah J. Johnson, MA, BS, Department of Pediatrics, at Penn State College of Medicine in Hershey, Pa., et al, recently published a study in the Journal of the American Medical Association (JAMA) Pediatrics that evaluated the ability of salivary microRNA to identify concussion in children. The salivary test of microRNA levels, Johnson and colleagues argued, does accurately identify the “duration and character of concussion symptoms.” According to the researchers, the test demonstrated high prognostic potential as a “toolset for facilitating concussion management” and may provide an additional biomarker source for use in clinical laboratory testing.
MicroRNA Offers New Biomarkers for Concussion Diagnosis
The study tested the saliva of 52 adolescents with a clinical diagnosis of mild traumatic brain injury in the form of concussion for specific microRNA expressions. Researchers identified five microRNA molecules which “accurately identify” patients with concussion symptoms. Three of those molecules served to diagnose specific symptoms of headache, fatigue, and memory difficulties up to one month after injury with low false detection rates. Because these microRNA molecules are not specific to children, could the test maintain diagnostic accuracy for patients of all ages?
William P. Meehan III, MD, with the Micheli Center for Sports Injury Prevention at Boston General Hospital, and Rebekah Mannix, MD, MPH, with the Brain Injury Center at Boston Children’s Hospital wrote an editorial responding to the original research article stating that “the use of salivary microRNA in this study is both novel and clinically relevant.” Adding that “using this salivary microRNA panel to diagnose and manage concussions could be a major advancement to the field.”
Meehan and Mannix also remarked on the speed and relative ease of obtaining saliva samples, stating that “salivary microRNAs could also offer insights into the underlying biological mechanisms of injuries, potentially identifying specific targets to modify disease.”
More Accurate than Current Concussion Diagnosis Tools
There has been a marked interest in microRNA analysis and testing in recent years. MicroRNA analysis and testing has found use in cancer prognosis and personalized medicine that help predict responses to specific treatments for individual patients with a variety of chronic diseases. The news that microRNA can be used to predict concussion and duration of symptoms further solidifies the role microRNA may play in medical laboratory testing in the near future.
In an interview with CNN, Steve Hicks, MD, PhD, senior author of the JAMA Pediatrics research article and Assistant Professor of Pediatrics at Penn State College of Medicine, reported that the salivary microRNA test predicted concussion with 85% accuracy in comparison to current clinical survey measures, which are “approximately 65% accurate.” Hicks added that “the technology required to measure saliva RNA is already employed in medicine” as a common means of testing for upper respiratory viruses and that “modifying this approach for patients with concussions could potentially provide a rapid, objective tool for managing brain injury.”
Currently the Standard Concussion Assessment Tool, Third Edition (SCAT 3), which includes a series of cognitive and physical tests, is used on sports sidelines to detect concussion symptoms. Hicks notes that one problem with SCAT 3 is that “an athlete may have a concussion even if [his or her] score is ‘normal.’” Therefore, the microRNA saliva test could provide objective evidence of concussion in patients SCAT 3 fails to accurately diagnose.

Steven D. Hicks, MD, PhD (above), led the research team that studied the use of microRNA in saliva, rather than in blood, as a biomarker to identify concussions symptoms in children, and determine how long effects of the injury might last. (Photo copyright: Penn State Health.)
Too Early to Know How Helpful the Test May Be?
In the same CNN interview, Neurologist Jeffery Kutcher, MD, head of the Sports Neurology Clinic at The Core Institute in Brighton, Mich., stated that the Penn State study’s findings were “promising” and that “work like this is important because it does provide potential for tests that can be helpful in the clinical setting.” Kutcher cautioned however, that it was “too early to know what this type of tool can do for us.”
In an NPR article, Manish Bhomia, M.Eng., PhD, a brain injury researcher with the Uniformed Services University of the Health Sciences commented that “a saliva test could greatly improve care for young people who don’t have obvious symptoms of a concussion.” Bhomia stated that “micro-RNAs offer a promising way to assess concussions in adults as well as children,” but he is wary to laud saliva tests as the best method of measuring relevant microRNA molecules. Bhomia states that blood samples “which tend to contain greater numbers of the genetic fragments” are perhaps a better option.
Hicks disagrees. In an article from Penn State News, Hicks stated that the novel aspect of this study was that it focused on microRNA levels “in saliva rather than blood.” Thus, a test based on saliva, rather than a phlebotomy stick or more invasive blood testing, requires no need for venous blood.
“The ultimate goal is to be able to objectively identify that a concussion has happened and then predict how long the symptoms will go on for,” Hicks noted in the Penn State News article. “Then, we can use that knowledge to improve the care that we provide for children who have concussions, either by starting medicine earlier or holding them out of activities for longer.”
Quadrant Biosciences, a biotech company in Syracuse, N.Y., that helped fund the study, is hoping to “bring a saliva test for concussion to market in the next 12 to 24 months,” according to Hicks in his CNN interview. If development proceeds as planned, the saliva test could prove a “game changer” for sports medicine diagnostics and possibly open new avenues for related microRNA in clinical laboratory testing.
—Amanda Warren
Related Information:
Prolonged Concussion Symptoms Identifiable by Salivary MicroRNA
Association of Salivary MicroRNA Changes with Prolonged Concussion Symptoms
Promise of Salivary MicroRNA for Assessing Concussion
Spit Test May Diagnose, Predict Duration of Concussion in Kids
Molecules in Spit May Be Able to Diagnose and Predict Length of Concussions
Spit Test May Help Reveal Concussion Severity
Meet the Clinical Pathology Laboratory on the Palm of a Hand: Japanese Researchers Announce A Point-Of-Care Testing Device That Detects MicroRNA in 20 Minutes
With Launch of RNAcentral Database, Pathologists Now Have Unprecedented Access to RNA Data
Feb 2, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
Research goal was to isolate circulating tumor cells in venipuncture samples with improved purity compared to standard spiral chips
Many research teams are pursuing the goal of creating assays that detect circulating tumor cells (CTCs) that would allow earlier and more accurate diagnosis of cancer. Now comes news of a unique technology developed at the University of Michigan (U-M) Ann Arbor that showed promised in an early study.
The method of using CTCs to diagnose cancer in patients, while further analyzing specific characteristics of a given cancer case, shows promise as an innovative tool for clinical laboratories and oncologists. However, current approaches face challenges when it comes to proving accuracy and establishing thresholds that might indicate the need for further action.
Researchers at U-M believe they may have solved that problem. They created “Labyrinth,” a “label-free microfluidic device” that condenses 637mm of channels—including 11 loops and 56 corners—onto a 500μm-wide chip that uses inertia and Dean flow to separate white blood cells and CTCs from venipuncture samples at rates as high as 2.5ml per minute. These results improve upon the traditional spiral chip design.
Publishing their findings in Cell Systems, first author of the study Eric Lin, PhD, noted, “With the recent advances in tools for genomic characterization, it is more compelling than ever to look at the tumor heterogeneity to understand tumor progression and resistance to therapies. The Labyrinth device enabled high yields of CTCs without the bias induced by antibody-based selection, allowing the identification of true biological tumor heterogeneity.”
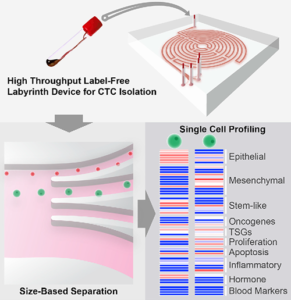
The graphic above, taken from the University of Michigan study, demonstrates the “High-throughput and label-free Labyrinth device that enables single CTC isolation and gene expression characterization.” According to the researchers, “Labyrinth offers a cell-surface marker-independent single-cell isolation platform to study heterogeneous CTC subpopulations.” The U-M study shows promise in creating tools for oncologist and clinical laboratory cancer treatment. (Image copyright: University of Michigan/Cell Systems.)
Challenges in the Isolation of CTCs
The Labyrinth chip is not the first device to assist in isolating CTCs. The U-M study notes that while immune-affinity capture is a validated approach to prognosis, therapeutic monitoring and molecular diagnostics, it does not work with all cancer cases. The researchers also note the method creates challenges in single-cell analysis later.
Existing label-free methods of isolation, such as deterministic lateral displacement, microfluidic flow fractionation, and acoustic-based separation, avoid these concerns but face issues of their own. The researchers noted, “Issues encountered with these approaches include pore clogging, high-pressure drop, pre-fixation to prevent CTC loss, low throughput, and excessive non-specific cell retention.”
The researchers further clarified that a major factor separating the Labyrinth chip from other methods is the ability to identify CTC subpopulations without the need for manual selection based on positive or negative protein expression. Thus, improving the ability to conduct further single-cell analysis from the results. Testing of the Labyrinth chip involved a variety of cancer cell lines, including:
· Human breast (MCF-7);
· Pancreatic (PANC-1);
· Prostate (PC-3); and,
· Lung (H1650).
And while standard spiral chips are already a common method for conducting size-based sorting, the purity of results is less than ideal with thousands of other cells remaining in the sample.
The researchers reported that the Labyrinth chip recovered 91.5% (plus or minus 0.9%) of cancer cells and removed 91.4% (plus or minus 3.3%) of white blood cells in a spiked buffer test.
“Bigger cells, like most cancer cells, focus pretty fast due to the curvature. But the smaller the cell is, the longer it takes to get focused,” Sunitha Nagrath, PhD, Associate Professor of Chemical Engineering and a lead developer of the Labyrinth chip, stated in a U-M news release. “The corners produce a mixing action that makes the smaller white blood cells come close to the equilibrium position much faster.”
Labyrinth also supports a series configuration of multiple chips. While testing two chips in series, researchers noted “a two-log improvement in tumor cell enrichment over the single Labyrinth.” They claim this is a higher purity than other label-free methods they studied, while adding only five minutes to processing times.

Sunitha Nagrath, PhD (above), is an Associate Professor of Chemical Engineering at the University of Michigan, and one of the lead developers of the Labyrinth chip. “You cannot put a box around these cells,” she noted in the U-M news release. “The markers for them are so complex, there is no one marker we could target for all these stages.” (Photo copyright: University of Michigan.)
Current Testing Using the Labyrinth Chip
The chip is already in use in a clinical trial for an aggressive form of breast cancer by Max Wicha, MD, Madeline and Sidney Forbes Professor of Oncology, Founding Director Emeritus, University of Michigan Comprehensive Cancer Center, and co-author of the Cell Systems study, who lead the study along with Nagrath.
The trial involves the attempted activation of adult system cells by blocking the signaling molecule interleukin-6. Wicha suspects the molecule enables cancer stem cells as well. “We think that this may be a way to monitor patients in clinical trials,” he said in the U-M news release. “Rather than just counting the cells, by capturing them, we can perform molecular analysis [to] know what we can target with treatments.”
The news release further highlights how this chip is specifically suited to such a task. As cancer stem cells transition from stem-like cells to more ordinary cell types, their gene expression shifts as well. This creates an issue when using conventional cell targeting. Nagrath notes this concern, stating, “The markers for [cancer stem cells] are so complex, there is no one marker we could target for all these stages.”
The Labyrinth chip shows potential for overcoming one of the biggest hurdles to leveraging CTCs to diagnose cancers and develop personalized therapies. Currently, the chip can output to Fluidigm, DEPArray by Silicon Biosystems, and RainDance Technologies’ RainDrop Digital PCR System.
The U-M researchers hope that future research will yield additional applications and compatible systems to further improve the ability for medical laboratories to use CTCs in the early detection and monitoring of cancer cases.
—Jon Stone
Related Information:
‘Labyrinth’ Chip Could Help Monitor Aggressive Cancer Stem Cells
High-throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells
Novel Labyrinth Chip Monitors Cancer Stem Cells in Clinical Trial
‘Labyrinth’ Device Sorts Cancer Cells from Healthy Blood
This Awesome Blood Labyrinth Is the Newest Method for Catching Cancer Cells
Labyrinth Chip Has the Potential to Lead to Customized Cancer Treatments
Jan 31, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Researchers are finding multiple approaches to metabolomic research and development involving disparate technology platforms and instrumentation
Human metabolome has been discovered to be a wealth of medical laboratory biomarkers for diagnosis, therapy, and patient monitoring. Because it can provide a dynamic phenotype of the human body, there are many potential clinical laboratory applications that could arise from metabolomics, the study of metabolites.
Researchers are discovering numerous ways the expanding field of metabolomics could transform the future of healthcare. However, to fully exploit the potential of human metabolome, developers must choose from various approaches to research.
“The metabolites we’re dealing with have vast differences in chemical properties, which means you need multi-platform approaches and various types of instrumentation,” James MacRae, PhD, Head of Metabolomics at the Francis Crick Institute in London, told Technology Networks. “We can either use an untargeted approach—trying to measure as much as possible, generating a metabolic profile—or else a more targeted approach where we are focusing on specific metabolites or pathways,” he added.
A multi-platform approach means different diagnostic technologies required to assess an individual’s various metabolomes, which, potentially, could result in multi-biomarker assays for medical laboratories.
Measuring All Metabolites in a Cell or Bio System
Metabolomics is the study of small molecules located within cells, biofluids, tissues, and organisms. These molecules are known as metabolites, and their functions within a biological system are cumulatively known as the metabolome.
Metabolomics, the study of metabolome, can render a real-time representation of the complete physiology of an organism by examining differences between biological samples based on their metabolite characteristics.
“Metabolomics is the attempt to measure all of the metabolites in a cell or bio system,” explained MacRae in the Technology Networks article. “You have tens of thousands of genes, of which tens of thousands will be expressed—and you also have the proteins expressed from them, which will then also be modified in different ways. And all of these things impact on a relatively small number of metabolites—in the thousands rather than the tens of thousands. Because of that, it’s a very sensitive output for the health or physiology of your sample.
“With that in mind, metabolomics has great potential for application in most, if not all, diseases—from diabetes, heart disease, cancer, HIV, autoimmune disease, parasitology, and host-pathogen interactions,” he added.

The graphic above is taken from a study published in the Journal of the American College of Cardiology (JACC). It notes, “State-of-the-art metabolomic technologies give us the ability to measure thousands of metabolites in biological fluids or biopsies, providing us with a metabolic fingerprint of individual patients. These metabolic profiles may serve as diagnostic and/or prognostic tools that have the potential to significantly alter the management of [chronic disease].” (Image and caption copyright:Journal of the American College of Cardiology.)
• Genomics: the study of DNA and genetic information within a cell;
• Proteomics: the large-scale study of proteins; and,
• Transcriptomics: the study of RNA and differences in mRNA expressions.
Researchers caution that metabolomics should be used in conjunction with other methods to analyze data for the most accurate results.
“Taking everything together—metabolic profiling, targeted assays, label incorporation and computational models, and also trying to associate all of this with proteomics and
genomics and transcriptomic data—that’s really what encapsulates both the power and also the challenges of metabolomics,” MacRae explained.
Metabolome in Precision Medicine
Metabolomics may also have the ability to help researchers and physicians fine-tune therapies to meet the specific needs of individual patients.
“We know we’re all very different and we don’t respond to drugs in the same way, so we could potentially use metabolomics to help select the best treatment for each individual,” Warwick Dunn, PhD, Senior Lecturer in Metabolomics at the University of Birmingham, Director of Mass Spectrometry, Phenome Center Birmingham, and, Co-Director, Birmingham Metabolomics Training Center, UK, told Technology Networks.
“Our genome is generally static and says what might happen in the future. And the metabolome at the other end is the opposite—very dynamic, saying what just happened or could be about the happen,” Dunn explained. “So, we could apply it to identify prognostic biomarkers, for example, to predict if someone is at greater risk of developing diabetes five to ten years from now. And if you know that, you can change their lifestyle or environment to try and prevent it.”
Metabolomics continues to tap the many diagnostic possibilities posed by the human metabolome. And, the resulting human biomarkers derived from the research could result in a rich new vein of medical laboratory assays.
—JP Schlingman
Related Information:
Metabolomics and Health: On the Cusp of a Revolution
‘Metabolomics’ Distinguishes Pancreatic Cancer from Pancreatitis
Using Metabolomics to Prevent Colon Cancer
Applications of Metabolomics
The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease
Metabolomics Takes Another Step Forward as Methodology for Clinical Laboratory Testing with Development of an Assay for the Diagnosis of Concussion










