Jul 25, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
While Apple recently debuted features to bring personal health records and protected health information to its mobile devices, Microsoft shuttered HealthVault in favor of focusing on AI-powered healthcare advances
As clinical laboratories and anatomic pathology groups know, lab testing data comprise more than 70% of the average patient’s health record. Thus, creating a universal platform on which consumers can share or review health information and medical histories with caregivers is a critical, yet elusive goal for most major tech companies, including tech giants Apple (Nasdaq:AAPL) and Microsoft (Nasdaq:MSFT).
Apple has big plans for patient health records and is working to bring protected health information (PHI) and healthcare advice to iPhones, iPads, and Apple Watch. Meanwhile, Microsoft is reducing its footprint in the mobile device healthcare market. Instead, it appears to be banking on its Artificial Intelligence (AI) platform. How these two diverging paths play out could have ramifications for the pathology and clinical laboratory industries.
HealthVault Insights versus AI versus Apple Health Mobile Apps
Launched in February 2017, Microsoft’s HealthVault Insights combined machine learning and AI with patients’ PHI and mobile activity tracking. The intent was to create an accessible, interactive platform for patients to monitor important health trends.
However, as of January 2018, Microsoft pulled the mobile app from Android, iOS, and Windows App stores. While summary information that draws on previously collected data is still available from the HealthVault website, new data and detailed insights are no longer available.
“We launched HealthVault Insights as a research project … with the goal of helping patients generate new insights about their health,” states Microsoft’s HealthVault Insights website. “Since then, we’ve learned a lot about how machine learning can be used to increase patient engagement and are now applying that knowledge to other projects.”
According to ZDNet, the closing of HealthVault Insights does not impact the Microsoft Health platform or the HealthVault patient-records system.
However, Microsoft’s shuttering of HealthVault Insights, and Google’s shuttering its Google Health platform in 2012, does seem to make Apple the last major tech company developing apps target at healthcare consumers designed to help them exchange private health information with caregivers through mobile devices. Dark Daily reported on Apple’s update earlier this year. (See, “Apple’s Update of Its Mobile Health App Consolidates Data from Multiple EHRs and Makes It Easier to Push Clinical Laboratory Data to Patients,” March 21, 2018.)
AI Will ‘Dramatically Transform Healthcare’
Shuttering HealthVault highlighted Microsoft’s shift away from consumer-facing health efforts and toward assisting medical laboratories, physicians, and research groups discover and implement treatments driving modern personalized medicine.
In a Microsoft blog post, Peter Lee, Corporate VP of Microsoft Healthcare, stated that Microsoft hopes its Healthcare NeXT platform will “dramatically transform healthcare, will deeply integrate Greenfield research and health technology product development, as well as establish a new model at Microsoft for strategic health industry partnerships.”
HealthVault Insights was one of several projects in Microsoft’s Healthcare NeXT initiative. Run by Microsoft’s AI and Research Group and partnering with major healthcare and research facilities across the country, other projects in the Healthcare NeXT initiative include:
Speaking with Business Insider, Lee noted that healthcare is becoming a “very large business” for Microsoft. “We don’t talk publicly about the dollars, but it’s large,” he concluded.
Microsoft’s EmpowerMD website states the eventual goal is to use the system to connect conversations with the growing trove of healthcare data available. “Our long-term vision is a learning system that incorporates data from longitudinal medical records, medical devices, genomics, population health, research papers, and more.”
AI a ‘Sleeping Giant for Healthcare’
“AI can be viewed as a sleeping giant for healthcare,” Eric Horvitz, PhD, Director of Microsoft Research Labs, told Nasdaq, when discussing Microsoft’s view of technology and healthcare. “AI methods show promise for multiple roles in healthcare. [This includes] inferring and alerting about hidden risks of potential adverse outcomes, selectively guiding attention, care, and interventional programs where [they are] most needed and reducing errors in hospitals.”
One such project involves a strategic partnership with the University of Pittsburg Medical Center (UPMC), which is a “$13-billion Pittsburgh-based system, comprising more than 25 hospitals, a three-million-member health plan, and 3,600 physicians, [that] will be a core partner in our efforts to improve healthcare delivery through a series of projects, beginning with a focus on transforming clinician empowerment and productivity,” according to Microsoft.

“Despite UPMC’s efforts to stay on the leading edge of technology, too often our clinicians and patients feel as though they’re serving the technology rather than the other way around. With Microsoft, we have a shared vision of empowering clinicians by reducing the burden of electronic paperwork and allowing the doctor to focus on the sacred doctor-patient relationship,” Steven D. Shapiro, MD (above), Chief Medical and Scientific Officer of UPMC and President of UPMC’s Health Services division, stated in the Microsoft blog. [Photo copyright: University of Pittsburg Medical Center.]
Medical laboratories and anatomic pathology groups may soon contribute health information to databases that one day will power AI systems. These trends highlight opportunities to both educate physicians on the tools available to utilize patient health data in an effective manner, and on new platforms that clinical laboratories could use to further streamline operations, reduce costs, and boost efficiency.
—Jon Stone
Related Information:
How Microsoft Is Using Advanced Technology in Healthcare
Microsoft Scrapping Personal Health Data App-Based Research Project
An Update on HealthVault Insights
How Microsoft’s Top Scientists Have Built a Big Business in Hacking Healthcare and Helped a Lot of People Along the Way
Microsoft Abandons Its Own HealthVault App: Is This Part of Something Larger?
Here’s How Microsoft Is Investing in AI
Microsoft Rolls Out More AI-Infused Healthcare Services, Software
Microsoft and Partners Combine the Cloud, AI, Research and Industry Expertise to Focus on Transforming Health Care
In Healthcare Push, Microsoft Launches Genomics Service on Azure Cloud
Apple’s Update of Its Mobile Health App Consolidates Data from Multiple EHRs and Makes It Easier to Push Clinical Laboratory Data to Patients
Jul 23, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Here is another example of how the diagnostic technologies used by anatomic pathology and clinical laboratories are evolving toward minute devices that deliver data wirelessly to electronic health records and healthcare databases for access by physicians and lab professionals
Anatomic pathologists and clinical laboratory managers keeping track of new implantable patient monitoring devices should find Abbott Laboratories’ (Abbott) recent technology release of great interest.
It’s an insertable cardiac monitor (ICM), also known as an implantable loop recorder, called Confirm Rx that, once implanted, can continuously monitor a patient’s heart rate and, using Bluetooth wireless technology, transmit collected data to physicians via a smartphone app. Abbott claims it is the “world’s first smartphone-compatible ICM.”
The laboratory’s website states, “The Confirm Rx ICM offers convenient, connected, and continuous monitoring for insight into your patients’ conditions and symptoms, including syncope, palpitations, and [atrial fibrillation] before or after ablation therapy and cryptogenic stroke—with fewer interruptions to their daily lives.”
Healthcare providers can implant Confirm Rx underneath the skin of a patient’s chest in just a few minutes using a simple insertion tool. The monitoring device is about the size of a small paper clip and has a two-year battery life. It provides physicians and their patients with an innovative way to identify and monitor abnormal heart rhythms or cardiac arrhythmias.
Transmitting ICM Data Using Smartphone App
Data collected by the Confirm Rx can be transmitted to a healthcare professional using the myMerlin for Confirm Rx smartphone app, developed by St. Jude Medical, which is now Abbott Laboratories (NYSE:ABT). No home-based transmitter, handheld activator, or additional equipment is required. However, individuals who use the Confirm Rx need to keep their smartphones within five feet of their bodies to ensure a continuous connection.
“Confirm Rx shows what we can do with cutting edge communication technology and the most advanced medical devices that provide new opportunities to improve patient care,” stated Avi Fischer, MD, Divisional VP Medical Affairs, and Medical Director for Cardiac Rhythm Management at Abbott, in a news release.
“By offering a device that uses Bluetooth wireless technology from the patient’s smartphone, we can help physicians easily and remotely diagnose potentially dangerous abnormal heart beats without requiring the patient to use a separate or cumbersome recording device,” Fischer concluded.
The myMerlin mobile app transmits its data to the Merlin.net Patient Care Network, where doctors and patients can simultaneously track symptoms, synchronize personal data, and view a transmission history.
“Prior to the evolution of this technology, the ability to monitor patients was really short-lived. But this allows the monitoring to continue and allows the patient to continue to live their lives unhindered,” noted Sean C. Beinart, MD, Cardiologist and Medical Director, Cardiovascular Research Washington Adventist Hospital, in an Abbott product release.

“Whenever you’re dealing with rhythm problems in the heart, being able to identify risk can sometimes mean the difference between life and death. The key is finding out what happened: What’s the cause of your symptoms?” Sean C. Beinart, MD (above), FACC, FHRS, a Cardiologist and Medical Director, Cardiovascular Research Washington Adventist Hospital, stated in an Abbott Laboratories video. Click on this link to watch the video. (Caption and video copyright: Abbott Laboratories.)
Peace of Mind through Technology
According to Abbott’s website, up to 7.1 million people in the United States are living with irregular heartbeats. The stroke risk for these individuals is five times higher than the risk for those without arrhythmias. When properly diagnosed, the condition is highly treatable.
“The stakes are very high,” Beinart stressed. “What the Confirm Rx does is it gives both me and my patient peace of mind that we’ve got them covered.”
Abbott obtained the Confirm Rx device when it acquired St. Jude Medical last year. The US Food and Drug Administration (FDA) approved the device in October of 2017. The first implantation of the Confirm Rx device took place in November.
The feedback received from users regarding the Confirm Rx has been positive and has helped increase Abbott’s electrophysiology sales by 16% during the last quarter of 2017, Fischer told CNBC.
“The types of patients have varied from younger to older, and [the devices] are being implanted for a variety of conditions,” Fischer stated. “Sometimes it’s to diagnose or to rule something out.”
As more implantable diagnostic monitoring/reporting devices are brought to the market, it is expected that some will be designed to measure some of the same biomarkers used in the assays performed by clinical laboratories. As that happens, clinical labs will have opportunities to serve medical facilities, physicians, and patients in new and more timely ways.
These implantable diagnostic devices may also give medical laboratories another way to be of value in the collection, storing, analysis, and reporting of the data obtained by the monitors, along with other lab tests doctors may order.
—JP Schlingman
Related Information:
Medicine Is Moving from Implantables to ‘Chipables’
Heart Monitor Links Patients to Doctors Using the One Thing They’re Unlikely to Forget Their Phones
Abbott Launches the First and Only Smartphone Compatible Insertable Cardiac Monitor in the U.S.
Heart Monitoring, Everywhere You Go
Confirm Rx Patient Brochure (PDF)
Jul 20, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Should greater attention be given to protein damage in chronic diseases such as Alzheimer’s and diabetes? One life scientist says “yes” and suggests changing how test developers view the cause of age-related and degenerative diseases
DNA and the human genome get plenty of media attention and are considered by many to be unlocking the secrets to health and long life. However, as clinical laboratory professionals know, DNA is just one component of the very complex organism that is a human being.
In fact, DNA, RNA, and proteins are all valid biomarkers for medical laboratory tests and, according to one life scientist, all three should get equal attention as to their role in curing disease and keeping people healthy.
Along with proteins and RNA, DNA is actually an “equal partner in the circle of life,” wrote David Grainger, PhD, CEO of Methuselah Health, in a Forbes opinion piece about what he calls the “cult of DNA-centricity” and its relative limitations.
Effects of Protein Damage
“Aging and age-related degenerative diseases are caused by protein damage rather than by DNA damage,” explained Grainger, a Life Scientist who studies the role proteins play in aging and disease. “DNA, like data, cannot by itself do anything. The data on your computer is powerless without apps to interpret it, screens and speakers to communicate it, keyboards and touchscreens to interact with it.”
“Similarly,” he continued, “the DNA sequence information (although it resides in a physical object—the DNA molecule—just as computer data resides on a hard disk) is powerless and ethereal until it is translated into proteins that can perform functions,” he points out.
According to Grainger, diseases such as cystic fibrosis and Duchenne Muscular Dystrophy may be associated with genetic mutation. However, other diseases take a different course and are more likely to develop due to protein damage, which he contends may strengthen in time, causing changes in cells or tissues and, eventually, age-related diseases.
“Alzheimer’s disease, diabetes, or autoimmunity often take decades to develop (even though your genome sequence has been the same since the day you were conceived); the insidious accumulation of the damaged protein may be very slow indeed,” he penned.
“But so strong is the cult of DNA-centricity that most scientists seem unwilling to challenge the fundamental assumption that the cause of late-onset diseases must lie somewhere in the genome,” Grainger concludes.
Shifting Focus from Genetics to Proteins
Besides being CEO of Methuselah Health, Grainger also is Co-Founder and Chief Scientific Advisor at Medicxi, a life sciences investment firm that backed Methuselah Health with $5 million in venture capital funding for research into disease treatments that focus on proteins in aging, reported Fierce CEO.
Methuselah Health, founded in 2015 in Cambridge, UK, with offices in the US, is reportedly using post-translational modifications for analysis of many different proteins.

“At Methuselah Health, we have shifted focus from the genetics—which tells you in an ideal world how your body would function—to the now: this is how your body functions now and this is what is going wrong with it. And that answer lies in the proteins,” stated Dr. David Grainger (above), CEO of Methuselah Health, in an interview with the UK’s New NHS Alliance. Click on this link to watch the full interview. [Photo and caption copyright: New NHS Alliance.]
This is how Methuselah Health analyzes damaged proteins using mass spectrometry, according to David Mosedale, PhD, Methuselah Health’s Chief Technology Officer, in the New NHS Alliance story:
- Protein samples from healthy individuals and people with diseases are used;
- Proteins from the samples are sliced into protein blocks and fed slowly into a mass spectrometer, which accurately weighs them;
- Scientists observe damage to individual blocks of proteins;
- Taking those blocks, proteins are reconstructed to ascertain which proteins have been damaged;
- Information is leveraged for discovery of drugs to target diseases.
Mass spectrometry is a powerful approach to protein sample identification, according to News-Medical.Net. It enables analysis of protein specificity and background contaminants. Interactions among proteins—with RNA or DNA—also are possible with mass spectrometry.
Methuselah Health’s scientists are particularly interested in the damaged proteins that have been around a while, which they call hyper-stable danger variants (HSDVs) and consider to be the foundation for development of age-related diseases, Grainger told WuXi AppTec.
“By applying the Methuselah platform, we can see the HSDVs and so understand which pathways we need to target to prevent disease,” he explained.
For clinical laboratories, pathologists, and their patients, work by Methuselah Health could accelerate the development of personalized medicine treatments for debilitating chronic diseases. Furthermore, it may compel more people to think of DNA as one of several components interacting that make up human bodies and not as the only game in diagnostics.
—Donna Marie Pocius
Related Information:
The Cult of DNA-Centricity
Methuselah Health CEO David Grainger Out to Aid Longevity
VIDEO: Methuselah Health, Addressing Diseases Associated with Aging
Understanding and Slowing the Human Aging Clock Via Protein Stability
Using Mass Spectrometry for Protein Complex Analysis
Jul 18, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory News, Laboratory Operations, Laboratory Testing, Management & Operations
MUSE microscope speeds up some anatomic pathology laboratory processes and removes exposure to toxic fixative chemicals
Because they handle tissue specimens, histotechnologists, anatomic pathologists, and hospital nurses are exposed to deadly chemicals such as formaldehyde, formalin, Xylene, and Toluene. The risks associated with these chemicals has been covered regularly by Dark Daily as recently as 2018 and as far back as 2011. (See, “Europe Implements New Anatomic Pathology Guidelines to Reduce Nurse Exposure to Formaldehyde and Other Toxic Histology Chemicals,” January 3, 2018; and, “Health of Pathology Laboratory Technicians at Risk from Common Solvents like Xylene and Toluene,” July 5, 2011.)
Now, scientists at the University of California at Davis (UC Davis) have developed a microscope that uses ultraviolet light (UV) to illuminate tissue samples. The UV microscope removes the need for traditional histology processes involved with preparation of tissue to produce conventional slides and makes it possible for anatomic pathologists to evaluate tissues without formalin fixation, according to a UC Davis news release.
“Here, we introduce a simple, non-destructive slide-free technique that, within minutes, provides high-resolution diagnostic histological images resembling those obtained from conventional hematoxylin and eosin histology,” the researchers wrote in their paper, published in Nature Biomedical Engineering.
High-resolution Biopsy Images in Minutes
The UV microscope relies on technology that UC Davis researchers dubbed MUSE, which stands for Microscopy with Ultraviolet Surface Excitation. According to the researchers, MUSE produces high-resolution images of biopsies and other fresh tissue samples that are ready for a pathologist’s review within minutes.
“MUSE eliminates any need for conventional tissue processing with formalin fixation, paraffin embedding, or thin-sectioning. It doesn’t require lasers, confocal, multiphoton, or optical coherence tomography instrumentation. And the simple technology makes it well-suited for deployment wherever biopsies are obtained and evaluated,” stated Richard Levenson, MD, MUSE Microscopy CEO, Professor, and Vice Chair for Strategic Technologies in the Department of Pathology and Laboratory Medicine at UC Davis, in the news release.
Ultraviolet microscopy is distinguished by its ability to magnify samples and enable views with greater resolution. This is due to the shorter wavelength of ultraviolet light, which improves image resolution beyond the diffraction limit of optical microscopes using normal white light, according to News Medical.
The unique ultraviolet light microscope tool may soon enable clinical laboratories and anatomic pathology groups to accurately report on biopsies to physicians and patients faster, for less money, and without exposure to deadly chemicals. This would be timely considering the pressure on the pathology industry to switch to value-based reimbursement from fee-for-service billing, and to embrace personalized medicine.
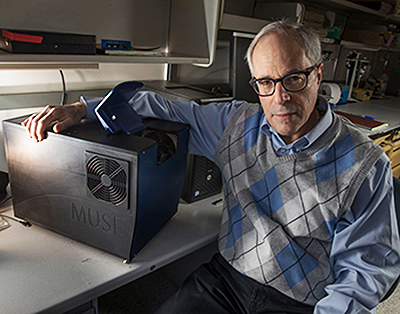
“It has become increasingly important to submit relevant portion of often tiny tissue samples for DNA and other molecular functional tests,” notes Richard Levenson, MD, MUSE Microscopy CEO, Professor, and Vice Chair for Strategic Technologies in the Department of Pathology and Laboratory Medicine at UC Davis, shown above with MUSE. “Making sure that the submitted material actually contains tumor in sufficient quantity is not always easy and sometimes just preparing conventional microscope slices can consume most of or even all of small specimens. MUSE is important because it quickly provides images from fresh tissue without exhausting the sample.” (Photo and caption copyright: UC Davis.)
MUSE is being commercialized and investors sought by MUSE Microscopy, Inc.
Traditional Microscopy is Time-Consuming, Hazardous, Expensive
Light microscopy, a time-honored technology, has been available to pathologists for more than 200 years. It is the cornerstone for cancer diagnostics and pathology, the UC Davis researchers acknowledged. But it requires time-consuming and expensive processes, which are especially glaring in a resource-challenged healthcare industry, they pointed out.
“Histological examination of tissues is central to the diagnosis and management of neoplasms and many other diseases. However, commonly used bright-field microscopy requires prior preparation of micrometer-thick tissue sections mounted on glass slides—a process that can require hours or days, contributes to cost, and delays access to critical information,” they wrote in their paper.
“MUSE promises to improve the speed and efficiency of patient care in both state-of-the art and low-resource settings, and to provide opportunities for rapid histology in research,” they continued.
No Histology Slide Preparation Needed
MUSE developers also called attention to the use of hazardous chemicals, such as formalin, in lab processes, which has been linked to cancers including myeloid leukemia, nasopharyngeal cancer, and sinonasal cancer, according to a National Academy of Sciences report. Still, more than 300 million slides are prepared in the US each year at a cost of several billion dollars to the healthcare industry, according to the MUSE Website.
MUSE, however, penetrates tissue samples by using ultraviolet light at short wavelengths—below the 300-nanometer range. The MUSE ultraviolet microscope can reach several microns-deep into tissues.
That’s enough, the researchers claim, to be comparable with the thickness of tissue slices anatomic pathologists use with traditional microscope slides. However, MUSE requires no conventional tissue processing associated with histology slides.
How Does it Work?
MUSE is comprised of an optical system with UV light-emitting diodes (LEDs), a UV compatible stage, and a conventional microscope. That’s according to Photonics Online, which described the process:
- “UV light at 280 nanometer spectral range illuminates about one square millimeter of specimen;
- “Surface is limited to a few nanometers deep to make high-contrast images possible;
- “Excitation light, at sub-300 nanometer spectral region, elicits bright emission from tissue specimens;
- “Specimens, which were stained with conventional florescent dyes, emit photons;
- “Photons are captured using glass-based microscope optics;
- “A Python programing language solution, with a graphics unit, converts MUSE images in real-time;
- “Images are comparable to the hematoxylin and eosin versions histologists and pathologists are accustomed to.”
The result, according the MUSE website, “is stunning detailed images conveying a degree of resolution, structure, and depth unachievable until now by any single technology.”
Other Alternative Histology Processes Under the Microscope
MUSE is not the only approach being studied that could create cellular images without sectioning tissue samples. Anatomic and histopathology laboratory leaders looking to differentiate their labs should keep watch on the development of MUSE and other alternatives to current histology methods, especially once these new devices become green-lighted by the Food and Drug Administration (FDA) for use in patient care.
—Donna Marie Pocius
Related Information:
Microscope That Uses Ultraviolet Instead of Visible Light Emerging as Powerful Diagnostic Tool
Microscope with Ultraviolet Surface Excitation for Rapid Slide-Free Histology
Ultraviolet Microscope to Dramatically Speed-up Lab Tests
What is Ultraviolet Microscopy?
Europe Implements New Anatomic Pathology Guidelines to Reduce Nurse Exposure to Formaldehyde and Other Toxic Histology Chemicals
National Academy of Sciences Confirms That Formaldehyde Can Cause Cancer in a Finding That Has Implications for Anatomic Pathology and Histology Laboratories
Health of Pathology Laboratory Technicians at Risk from Common Solvents like Xylene and Toluene
Jul 16, 2018 | Laboratory News, Laboratory Operations, Laboratory Testing, News From Dark Daily
This potential new source of diagnostic biomarkers could give clinical labs a new tool to diagnose disease earlier and with greater accuracy
Clinical laboratories may soon have a new “omics” in their toolkit and vocabulary. In addition to genomics and proteomics, anatomic pathologists could also be using “interactomics” to diagnose disease earlier and with increased accuracy.
At least that’s what researchers at ETH Zurich (ETH), an international university for technology and natural sciences, have concluded. They published the results of their study in Cell.
“Here, we present a chemoproteomic workflow for the systematic identification of metabolite-protein interactions directly in their native environments,” the researchers wrote. “Our data reveal functional and structural principles of chemical communication, shed light on the prevalence and mechanisms of enzyme promiscuity, and enable extraction of quantitative parameters of metabolite binding on a proteome-wide scale.”
Interactomics address interactions between proteins and small molecules, according to an article published in Technology Networks. The terms “interactomics” and “omics” were inspired by research that described, for the first time, the interactions and relationships of all proteins and metabolites (A.K.A, small molecules) in the whole proteome.
Medical laboratories and anatomic pathologists have long understood the interactions among proteins, or between proteins and DNA or RNA. However, metabolite interactions with packages of proteins are not as well known.
These new omics could eventually be an important source of diagnostic biomarkers. They may, one day, contribute to lower cost clinical laboratory testing for some diseases, as well.
Metabolite-Protein Interactions are Key to Cellular Processes
The ETH researchers were motivated to explore the interplay between small molecules and proteins because they have important responsibilities in the body. These cellular processes include:
“Metabolite-protein interactions control a variety of cellular processes, thereby playing a major role in maintaining cellular homeostasis. Metabolites comprise the largest fraction of molecules in cells. But our knowledge of the metabolite-protein interaction lags behind our understanding of protein-protein or protein-DNA interactomes,” the researchers wrote in Cell.
Leveraging Limited Proteolysis and Mass Spectrometry
The researchers used limited proteolysis (LiP) technology with mass spectrometry to discover metabolite-protein interactions. Results aside, experts pointed out that the LiP technology itself is significant.
“It is one of the few methods that enables the unbiased and proteome-wide profiling of protein conformational changes resulting from interaction of proteins with compounds,” stated a Biognosys blog post.
Biognosys, a proteomics company founded in 2008, was originally part of a lab at ETH Zurich.
The ETH team focused on the E. coli bacterial cell in particular and how its proteins and enzymes interact with metabolites.

“Although the metabolism of E. coli and associated molecules is already very well known, we succeeded in discovering many new interactions and the corresponding binding sites,” Paola Picotti, PhD, Professor of Molecular Systems Biology at ETH Zurich, who led the research, told Technology Networks. “The data that we produce with this technique will help to identify new regulatory mechanisms, unknown enzymes and new metabolic reactions in the cell,” she concluded. (Photo copyright: ETH Zurich.)
More than 1,000 New Interactions Discovered
The study progressed as follows, according to Technology Networks’ report:
- “Cellular fluid, containing proteins, was extracted from bacterial cells;
- “A metabolite was added to each sample;
- “The metabolite interacted with proteins;
- “Proteins were cut into smaller pieces by molecular scissors (A.K.A., CRISPR-Cas9);
- “Protein structure was altered when it interacted with a metabolite;
- “A different set of peptides emerged when the “molecular scissors” cut at different sites;
- “Pieces of samples were measured with a mass spectrometer;
- “Data were obtained, fed into a computer, and structural differences and changes were reconstructed;
- “1,650 different protein-metabolite interactions were found;
- “1,400 of those discovered were new.”
A Vast, Uncharted Metabolite-protein Interaction Network
The research is a major step forward in the body of knowledge about interactions between metabolites and proteins and how they affect cellular processes, according to Balázs Papp, PhD, Principal Investigator, Biological Research Center of the Hungarian Academy of Sciences.
“Strikingly, more than 80% of the reported interactions were novel and about one quarter of the measured proteome interacted with at least one of the 20 tested metabolites. This indicates that the metabolite-protein interaction network is vast and largely uncharted,” Papp stated in an ETH Zurich Faculty of 1000 online article.
According to Technology Networks, “Picotti has already patented the method. The ETH spin-off Biognosys is the exclusive license holder and is now using the method to test various drugs on behalf of pharmaceutical companies.”
The pharmaceutical industry is reportedly interested in the approach as a way to ascertain drug interactions with cellular proteins and their effectiveness in patient care.
The ETH Zurich study is compelling, especially as personalized medicine takes hold and more medical laboratories and anatomic pathology groups add molecular diagnostics to their capabilities.
—Donna Marie Pocius
Related Information:
The New “Omics”—Measuring Molecular Interactions
Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication
A New Study Maps Protein-Metabolite Interactions in an Unbiased Way
Cell Paper on Protein Metabolite Interactions Recommended in Faculty 1000 Twice
Jul 13, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Management & Operations, News From Dark Daily
Popularity of the pocket-sized gene-sequencing device continues to prove that DNA testing away from clinical laboratories in remote clinics and outlying field laboratories is not just possible, but in some cases preferable
Once again, Oxford Nanopore Technologies (ONT) is demonstrating how next-generation gene sequencing technology can make it cheaper, simpler, and faster to sequence without the need for big clinical laboratories. And its successful raising of $180 million to expand development worldwide shows the support it has with capital funding investors.
Dark Daily has repeatedly reported on the development of the UK-based company’s point-of-care DNA sequencer going back to 2011. Called MinION, we predicted in 2015, that once brought to market, the pocket-sized gene sequencing machine “could help achieve the NIH’s goal of $1,000 human genome sequencing and, in remote clinics and outbreak zones, shift testing away from medical laboratories.” (See Dark Daily, “Point-of-Care DNA Sequencer Inching Closer to Widespread Use as Beta-Testers Praise Oxford Technologies’ Pocketsize, Portable Nanopore Device,” November 4, 2015.)
Since then, MinION’s use worldwide “for a number of biological analysis techniques including de novo sequencing, targeted sequencing, metagenomics, epigenetics, and more” has only expanded, according to multiple sources and ONT’s website.
How Does MinION Work as a Gene Sequencer?
The MinION nanopore sequencing device weighs about 100 grams (less than four ounces), is about the size of a standard deck of cards, operates off a laptop USB plug, and can sequence genetic material in a matter of minutes.
To perform the nanopore sequencing, a strand of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) is pushed through small pores in a membrane. An ionic current is then applied to the material and voltage is implemented to measure any disruptions in the current. The resulting measurement represents an electrical signal that is converted to human-readable sequence.
“It’s like the ultimate barcode,” Gordon Sanghera, PhD, Chief Executive Officer at Oxford Nanopore, told BBC News.

Oxford Nanopore Technologies’ diminutive MinION gene-sequencing device has the capacity to directly recognize epigenetic markers that control gene activity and cellular processes involved in the onset and development of disease. Early detection of cancers, testing for birth defects and infectious diseases, and blood screening are possible future clinical laboratory applications for the MinION. Click on this link to watch video on MinION. (Photo copyright: Oxford Nanopore Technologies.)
Why is MinION Important?
One advantage to this technology is that it has the ability to sequence much longer strands of DNA when compared to existing technologies. The MinION can sequence over a million letters or bases, around 2% of a DNA strand or chromosome with 96% or above accuracy. The device can read remarkably long stretches of consecutive DNA letters. Readouts of several thousand letters are common and the record for the MinION is 882,000 consecutive DNA letters, Technology Review noted.
“One of the most important findings of this research was that, even though the human genome reference was completed or thought to have been completed a while ago, it still contains many missing pieces and we were able to close some of those gaps in the sequence by developing a new method for developing these extremely long reads using nanopore sequencing,” Nick Loman, PhD, Professor of Microbial Genomics and Bioinformatics at the School of Biosciences at the University of Birmingham, UK, told Pharmaphorum. Loman worked on research with Oxford Nanopore on nanopore sequencing.
“We’ve gone from a situation where you can only do genome sequencing for a huge amount of money in well-equipped labs to one where we can have genome sequencing literally in your pocket just like a mobile phone,” Loman told BBC News. “That gives us a really exciting opportunity to start having genome sequencing as a routine tool, perhaps something people can do in their own home.”
Using MinION in the Field
According to the Oxford Nanopore website, the MinION:
- Is pocket-sized and portable;
- Has up to 512 nanopore channels;
- Has a simple 10-minute sample preparation time;
- Allows real-time analysis for rapid and efficient results; and,
- Is adaptable to direct DNA or RNA sequencing.
The MinION Starter Pack is available for purchase on the company’s website with prices starting at $1,000. The kit includes:
- The MinION device;
- Flow cells;
- Sequencing kits;
- Wash kits; and,
- MinION community support.
Researchers at The Kinghorn Center for Clinical Genomics at the Garvan Institute of Medical Research in Darlinghurst, Australia, are currently using the MinION for research purposes.

Members of the Zebra Project (above), an international group of scientists, used Oxford Nanopore Technologies’ MinION to sequence genomes during epidemics in Latin America. With just a laptop computer for power, MinION can run complex gene-sequencing and achieve superior results than other similar technologies. It is in use worldwide bringing clinical laboratory testing to patients in remote, outlying locations. (Photo copyright: Ricardo Funari.)
“I think it’s really expanding the arsenal of tools we have to peer into cell biology and the root causes of cancer and various diseases,” Dr. Martin Smith, Head of Genomic Technologies at the center, told Australian Financial Review. “It’s really just starting to open the lid off the jar and peer more deeply into the genomics of the cell.”
Dr. Sanghera hopes the gadget could be utilized in the future to identify common infections at home and help consumers avoid unnecessary trips to doctors, clinics, and hospitals, and avert the misuse and overuse of prescription medications. He also feels MinION has applications outside the healthcare industry, such as detecting the presence of harmful microbes in food and water supplies.
As gadgets like MinION become more popular, the potential to move DNA sequencing closer to the patient (and out of the core lab) has implications for clinical laboratories and anatomic pathology groups. However, core labs would still be a preferred source to collect the raw data, store that data, then do the annotation of the DNA sequences and report the findings to the referring physician.
—JP Schlingman
Related Information:
How Knowing Your Genetic Code Could Lengthen Your Life
Genome in the Palm of Your Hand
Molecular Machines and the Place of Physics in the Biology Curriculum
Oxford Nanopore’s Hand-Held DNA Analyzer Has Traveled the World
Hostplus Sinks $27m Into Hand-held DNA Sequencing Firm Oxford Nanopore
GIC, Others Invest £100m In Hand-held DNA Sequencing firm Oxford Nanopore
Handheld Device Sequences Human Genome
Breakthrough Leads to Sequencing of a Human Genome Using a Pocket-sized Device
Oxford Nanopore’s Tech Reaches Genome Sequencing Landmark
Point-of-Care DNA Sequencer Inching Closer to Widespread Use as Beta-Testers Praise Oxford Technologies’ Pocketsize, Portable Nanopore Device
$900 Point-of-Care DNA Nanopore Sequencer May Hit Market in Next 12 Months
Is Whole-genome Sequencing Reaching a Tipping Point for Clinical Pathology Laboratories?










