Oct 10, 2018 | Coding, Billing, and Collections, Compliance, Legal, and Malpractice, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
Protecting patient privacy is of critical importance, and yet researchers reidentified data using only a few additional data points, casting doubt on the effectiveness of existing federally required data security methods and sharing protocols
Clinical laboratories and anatomic pathologists know the data generated by their diagnostics and testing services constitute most of a patient’s personal health record (PHR). They also know federal law requires them to secure their patients’ protected health information (PHI) and any threat to the security of that data endangers medical laboratories and healthcare practices as well.
Therefore, recent coverage in The Guardian which reported on how easily so-called “deidentified data” can be reidentified with just a few additional data points should be of particular interest to clinical laboratory and health network managers and stakeholders.
Risky Balance Between Data Sharing and Privacy
In December 2017, University of Melbourne (UM) researchers, Chris Culnane, PhD, Benjamin Rubinstein, and Vanessa Teague, PhD, published a report with the Cornell University Library detailing how they reidentified data listed in an open dataset of Australian medical billing records.
“We found that patients can be re-identified, without decryption, through a process of linking the unencrypted parts of the record with known information about the individual such as medical procedures and year of birth,” Culnane stated in a UM news release. “This shows the surprising ease with which de-identification can fail, highlighting the risky balance between data sharing and privacy.”
In a similar study published in Scientific Reports, Yves-Alexandre de Montjoye, PhD, a computation private researcher, used location data on 1.5 million people from a mobile phone dataset collected over 15 months to identify 95% of the people in an anonymized dataset using four unique data points. With just two unique data points, he could identify 50% of the people in the dataset.
“Location data is a fingerprint. It’s a piece of information that’s likely to exist across a broad range of data sets and could potentially be used as a global identifier,” Montjoye told The Guardian.
The problem is exacerbated by the fact that everything we do online these days generates data—much of it open to the public. “If you want to be a functioning member of society, you have no ability to restrict the amount of data that’s being vacuumed out of you to a meaningful level,” Chris Vickery, a security researcher and Director of Cyber Risk Research at UpGuard, told The Guardian.
This privacy vulnerability isn’t restricted to just users of the Internet and social media. In 2013, Latanya Sweeney, PhD, Professor and Director at Harvard’s Data Privacy Lab, performed similar analysis on approximately 579 participants in the Personal Genome Project who provided their zip code, date of birth, and gender to be included in the dataset. Of those analyzed, she named 42% of the individuals. Personal Genome Project later confirmed 97% of her submitted names according to Forbes.

In testimony before the Privacy and Integrity Advisory Committee of the Department of Homeland Security (DHS), Latanya Sweeney, PhD (above), Professor and Director at Harvard’s Data Privacy Lab stated, “One problem is that people don’t understand what makes data unique or identifiable. For example, in 1997 I was able to show how medical information that had all explicit identifiers, such as name, address and Social Security number removed could be reidentified using publicly available population registers (e.g., a voter list). In this particular example, I was able to show how the medical record of William Weld, the Governor of Massachusetts of the time, could be reidentified using only his date of birth, gender, and ZIP. In fact, 87% of the population of the United States is uniquely identified by date of birth (e.g., month, day, and year), gender, and their 5-digit ZIP codes. The point is that data that may look anonymous is not necessarily anonymous. Scientific assessment is needed.” (Photo copyright: US Department of Health and Human Services.)
These studies reveal that—regardless of attempts to create security standards—such as the Privacy Rule in the Health Insurance Portability and Accountability Act of 1996 (HIPAA)—the sheer amount of available data on the Internet makes it relatively easy to reidentify data that has been deidentified.
The Future of Privacy in Big Data
“Open publication of deidentified records like health, census, tax or Centrelink data is bound to fail, as it is trying to achieve two inconsistent aims: the protection of individual privacy and publication of detailed individual records,” Dr. Teague noted in the UM news release. “We need a much more controlled release in a secure research environment, as well as the ability to provide patients greater control and visibility over their data.”
While studies are mounting to show how vulnerable deidentified information might be, there’s little in the way of movement to fix the issue. Nevertheless, clinical laboratories should consider carefully any decision to sell anonymized (AKA, blinded) patient data for data mining purposes. The data may still contain enough identifying information to be used inappropriately. (See Dark Daily, “Coverage of Alexion Investigation Highlights the Risk to Clinical Laboratories That Sell Blinded Medical Data,” June 21, 2017.)
Should regulators and governments address the issue, clinical laboratories and healthcare providers could find more stringent regulations on the sharing of data—both identified and deidentified—and increased liability and responsibility regarding its governance and safekeeping.
Until then, any healthcare professional or researcher should consider the implications of deidentification—both to patients and businesses—should people use the data shared in unexpected and potentially malicious ways.
—Jon Stone
Related Information:
‘Data Is a Fingerprint’: Why You Aren’t as Anonymous as You Think Online
Research Reveals De-Identified Patient Data Can Be Re-Identified
Health Data in an Open World
The Simple Process of Re-Identifying Patients in Public Health Records
Harvard Professor Re-Identifies Anonymous Volunteers in DNA Study
How Someone Can Re-Identify Your Medical Records
Trading in Medical Data: Is this a Headache or An Opportunity for Pathologists and Clinical Laboratories
Coverage of Alexion Investigation Highlights the Risk to Clinical Laboratories That Sell Blinded Medical Data
Oct 8, 2018 | Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Both health systems will use their EHRs to track genetic testing data and plan to bring genetic data to primary care physicians
Clinical laboratories and pathology groups face a big challenge in how to get appropriate genetic and molecular data into electronic health record (EHR) systems in ways that are helpful for physicians. Precision medicine faces many barriers and this is one of the biggest. Aside from the sheer enormity of the data, there’s the question of making it useful and accessible for patient care. Thus, when two major healthcare systems resolve to accomplish this with their EHRs, laboratory managers and pathologists should take notice.
NorthShore University HealthSystem in Illinois and Geisinger Health System in Pennsylvania and New Jersey are working to make genetic testing part of primary care. And both reached similar conclusions regarding the best way for primary care physicians to make use of the information.
One area of common interest is pharmacogenomics.
At NorthShore, two genetic testing programs—MedClueRx and the Genetic and Wellness Assessment—provide doctors with more information about how their patients metabolize certain drugs and whether or not their medical and family histories suggest they need further, more specific genetic testing.
“We’re not trying to make all of our primary care physicians into genomic experts. That is a difficult strategy that really isn’t scalable. But we’re giving them enough tools to help them feel comfortable,” Peter Hulick, MD, Director of the Center for Personalized Medicine at NorthShore, told Healthcare IT News.
Conversely, Geisinger has made genomic testing an automated part of primary care. When patients visit their primary care physicians, they are asked to sign a release and undergo whole genome sequencing. An article in For the Record describes Geisinger’s program:
“The American College of Medical Genetics and Genomics classifies 59 genes as clinically actionable, with an additional 21 others recommended by Geisinger. If a pathogenic or likely pathogenic variant is found in one of those 80 genes, the patient and the primary care provider are notified.”
William Andrew Faucett (left) is Director of Policy and Education, Office of the Chief Scientific Officer at Geisinger Health; and Peter Hulick, MD (right), is Director of the Center for Personalized Medicine at NorthShore University HealthSystem. Both are leading programs at their respective healthcare networks to improve precision medicine and primary care by including genetic testing data and accessibility to it in their patients’ EHRs. (Photo copyrights: Geisinger/NorthShore University HealthSystem.)
The EHR as the Way to Access Genetic Test Results
Both NorthShore and Geisinger selected their EHRs for making important genetic information accessible to primary care physicians, as well as an avenue for tracking that information over time.
Hulick told Healthcare IT News that NorthShore decided to make small changes to their existing Epic EHR that would enable seemingly simple but actually complex actions to take place. For example, tracking the results of a genetic test within the EHR. According to Hulick, making the genetic test results trackable creates a “variant repository,” also known as a Clinical Data Repository.
“Once you have that, you can start to link it to other information that’s known about the patient: family history status, etc.,” he explained. “And you can start to build an infrastructure around it and use some of the tools for clinical decision support that are used in other areas: drug/drug interactions, reminders for flu vaccinations, and you can start to build on those decision support tools but apply them to genomics.”
Like NorthShore, Geisinger is also using its EHR to make genetic testing information available to primary care physician when a problem variant is identified. They use EHR products from both Epic and Cerner and are working with both companies to streamline and simplify the processes related to genetic testing. When a potentially problematic variant is found, it is listed in the EHR’s problem list, similar to other health issues.
Geisinger has developed a reporting system called GenomeCOMPASS, which notifies patients of their results and provides related information. It also enables patients to connect with a geneticist. GenomeCOMPASS has a physician-facing side where primary care doctors receive the results and have access to more information.
Andrew Faucett, Senior Investigator (Professor) and Director of Policy and Education, Office of the Chief Scientific Officer at Geisinger, compares the interpretation of genetic testing to any other kind of medical testing. “If a patient gets an MRI, the primary care physicians doesn’t interpret it—the radiologist does,” adding, “Doctors want to help patients follow the recommendations of the experts,” he told For the Record.
The Unknown Factor
Even though researchers regularly make new discoveries in genomics, physicians practicing today have had little, if any, training on how to incorporate genetics into their patients’ care. Combine that lack of knowledge and training with the current lack of EHR interoperability and the challenges in using genetic testing for precision medicine multiply to a staggering degree.
One thing that is certain: the scientific community will continue to gather knowledge that can be applied to improving the health of patients. Medical pathology laboratories will play a critical role in both testing and helping ensure results are useful and accessible, now and in the future.
—Dava Stewart
Related Information:
Introducing “Genomics and Precision Health”
How NorthShore Tweaked Its Epic EHR to Put Precision Medicine into Routine Clinical Workflows
Precise, Purposeful Health Care
Next-Generation Laboratory Information Management Systems Will Deliver Medical Laboratory Test Results and Patient Data to Point of Care, Improving Outcomes, Efficiency, and Revenue
Oct 5, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Pathology, Laboratory Testing, Management & Operations
New dichromatic color scale developed by scientists at the Pacific Northwest National Laboratory could play a role in how slides are stained and how software color-codes digital pathology images in ways that make it easier for human eyes to recognize structures and features of interest
Clinical laboratories, anatomic pathologists, and other specialized diagnostics providers play an essential role in precision medicine. Imagine, however, performing surgical pathology analysis on slides using displays that cannot recreate—or worse, inaccurately display—a range of colors used in the image being analyzed.
As many as 8% of men and 0.5% of women of Northern European ancestry already experience issues discerning colors in the interfaces, information, and world around them due to red-green color blindness according to the National Eye Institute. This can lead to potential for misreadings and medical errors.
Now, research from Pacific Northwest National Laboratory (PNNL) holds the potential to establish a standard colormap that eliminates the impact of red-green colorblindness on visuals. Surgical pathologists, for example, spend much of their days viewing slides and/or digital pathology images. Thus, any new method of illustrating/coloring/highlighting features of interest could eventually prove to be a useful innovation in the specialty of anatomic pathology.
In completing their research, the PNNL scientists created an open-source tool called Cmaputil that other researchers can use. Could it enable clinicians and laboratory workers to improve the visibility of critical elements in samples, slides, and other visual data formats used daily at medical laboratories and anatomic pathology groups?
PLOS One published details about the development of the colormap and its potential scientific applications in August.
PNNL’s Cividis Color Scale: A Better Alternative to Rainbow Color Scales?
While the typical rainbow color map draws attention to a chart or image, it is not particularly great at conveying information—especially if the reader is color vision deficient (CVD) or color blind. Yet, despite this, rainbow scales are common in everything from local weather reports and news stories to medical images and medical studies.

Jamie Nuñez, lead author of the PLOS One study and a chemical and biological data analyst at PNNL, told Scientific American, “People like to use rainbow because it catches the eye. But once the eye actually gets there, and people are trying to figure out what’s actually going on inside of the image, that’s kind of where it falls apart.” (Photo copyright: University of Washington.)
PNNL scientists started with the viridis colormap due to “its wide range of colors” and “overall sharpness when overlaid with complex images.” They created an open-source software tool capable of taking existing color scales and simulating the visual effect of red-green color blindness using a mathematical model of human sight. Their software adjusts the scale so that color and brightness vary at a steady rate.
Their adaptions resulted in what they call the “cividis colormap.” It is a blue and yellow scale that provides an accurate change in hue and luminance when compared to changes in the data set. Researchers noted that, to their knowledge, this is the first study to mathematically optimize a colormap specifically for viewing by both those with CVD and those with normal vision.
“Here, we present an example CVD-optimized colormap created with this module that is optimized for viewing by those without a CVD as well as those with red-green colorblindness. This colormap, cividis, enables nearly-identical visual-data interpretation to both groups, is perceptually uniform in hue and brightness, and increases in brightness linearly,” the researchers noted in the PLOS One study.
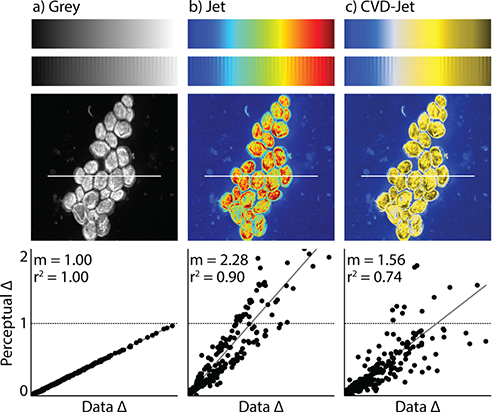
Example above is of a misleading colormap, taken from the PNNL/PLOS One study. An image of yeast cells is shown in gray scale (left), with a rainbow color scale (middle) and as a person with red-green color blindness sees the rainbow image (right). (Photo/caption copyright: Nuñez JR, Anderton CR, Renslow RS (2018) PLoS ONE 13(7): e0199239/Scientific American.)
The PNNL researchers report that the colormap will soon be ready in a number of tools, including:
According to Scientific American, cividis will be added to the color-scale libraries of roughly a dozen software packages.
“While it may take some time for the full scientific community to both be aware of the need to choose appropriate colormaps and agree on preferred colormaps,” PNNL researchers note, “we hope the code we provide here can help with this transition by allowing others to experiment with the different aspects of colormap design and see how the various characteristics of a colormap affect its interpretation.”
They are concerned that the changing color spaces on future displays may make current colormaps and standards obsolete, as they display colors outside the standard sRGB color space. However, the researchers also note that any change to color spaces could result in an increase in color availability and allow cmaputil to create better-optimized color schemes.
How Cividis and Similar Approaches Might Impact Pathology
While the technology was developed with mass spectrometry and fluid flow analysis in mind, it could prove useful for medical laboratories and specialized diagnostics providers as well—in particular, anatomic pathology and surgical pathology labs.
Coverage of a presentation at the 2011 IEEE Information Visualization Conference by Phys.org highlights a similar concept for diagnosing heart disease. By taking 3D representations of arteries using a rainbow colormap and converting them to 2D projections using a dichromatic black to red colormap, Harvard researchers found their HemoVis tool increased diagnostic accuracy from 39% to 91% in their study.
Technologies and techniques designed for scientific applications often find use in healthcare environments. For anatomic and surgical pathologist and other diagnostics providers, the research from PNNL shows promise for adapting the latest data visualization trends to further improve accuracy, efficiency, and accessibility of medical images, samples, and other complex images used daily in the process of diagnosing disease.
—Jon Stone
Related Information:
End of the Rainbow? New Map Scale Is More Readable by People Who Are Color Blind
Optimizing Colormaps with Consideration for Color Vision Deficiency to Enable Accurate Interpretation of Scientific Data
Evaluation of Artery Visualizations for Heart Disease Diagnosis
NanoSIMS for Biological Applications: Current Practices and Analyses
New Color Scale Makes Data Visualizations Easier for Colorblind People to Read
The End of the Rainbow? Color Schemes for Improved Data Graphics
Time to Replace ‘Rainbow Color Scale’ for Data Visualization?
How a New Color Scale for Scientific Models Could Improve Healthcare
To Diagnose Heart Disease, Visualization Experts Recommend a Simpler Approach
Oct 3, 2018 | Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
Next step is to design Web portal offering low-cost ‘polygenic risk score’ to people willing to upload genetic data received from DNA testing companies such as 23andMe
Pathologists and other medical professionals have long predicted that multi-gene diagnostics tests which examine thousands of specific gene sequences might one day hold the key to assessing disease risk, diagnosing diseases, and guiding precision medicine treatment decisions. Now, a research team from the Broad Institute, Massachusetts General Hospital (MGH) and Harvard Medical School have brought that prediction closer to reality.
Their study, published last month in Nature Genetics, found that a genome analysis called polygenic risk scoring can identify individuals with a high risk of developing one of five potentially deadly diseases:
- Coronary artery disease;
- Atrial fibrillation;
- Type 2 diabetes;
- Inflammatory bowel disease; and,
- Breast cancer.
Polygenic Scoring Predicts Risk of Disease Among General Population
To date, most genetic testing has been “single gene,” focusing on rare mutations in specific genes such as those causing sickle cell disease or cystic fibrosis. This latest research indicates that polygenic predictors could be used to discover heightened risk factors in a much larger portion of the general population, enabling early interventions to prevent disease before other warning signs appear. The ultimate goal of precision medicine.
“We’ve known for long time that there are people out there at high risk for disease based just on their overall genetic variation,” senior author Sekar Kathiresan, MD, co-Director of the Medical and Population Genetics Program at the Broad Institute, and Director, Center for Genomic Medicine at Massachusetts General Hospital, said in a Broad Institute news release. “Now, we’re able to measure that risk using genomic data in a meaningful way. From a public health perspective, we need to identify these higher-risk segments of the population, so we can provide appropriate care.”

“What I foresee is in five years, each person will know this risk number—this ‘polygenic risk score’—similar to the way each person knows his or her cholesterol,” Sekar Kathiresan, MD (above), Co-Director of the Medical and Population Genetics Program at the Broad Institute, and Director, Center for Genomic Medicine at Massachusetts General Hospital, told the Associated Press (AP). He went on to say a high-risk score could lead to people taking other steps to lower their overall risk for specific diseases, while a low-risk score “doesn’t give you a free pass” since an unhealthy lifestyle can lead to disease as well. (Photo copyright: Massachusetts General Hospital.)
The researchers conducted the study using data from more than 400,000 individuals in the United Kingdom Biobank. They created a risk score for coronary artery disease by looking for 6.6 million single-letter genetic changes that are more prevalent in people who have had early heart attacks. Of the individuals in the UK Biobank dataset, 8% were more than three times as likely to develop the disease compared to everyone else, based on their genetic variation.
In absolute terms, only 0.8% of individuals with the very lowest polygenic risk scores had coronary artery disease, compared to 11% for people with the highest scores, the Broad Institute news release stated.
“The results should be eye-opening for cardiologists,” Charles C. Hong, MD, PhD, Director of Cardiovascular Research at the University of Maryland School of Medicine, told the AP. “The only disappointment is that this score applies only to those with European ancestry, so I wonder if similar scores are in the works for the large majority of the world population that is not white.”
In its news release, the Broad Institute noted the need for additional studies to “optimize the algorithms for other ethnic groups.”
The Broad Institute’s results suggest, however, that as many as 25 million people in the United States may be at more than triple the normal risk for coronary artery disease. And millions more may be at similar elevated risk for the other conditions, based on genetic variations alone.
Reanalyzing Data from DNA Testing Companies
The researchers are building a website that would enable users to receive a low-cost polygenic risk score—such as calculating inherited risk score for many common diseases—by reanalyzing data users previously receive from DNA testing companies such as 23andMe.
Kathiresan told Forbes his goal is for the 17 million people who have used genotyping services to submit their data to the web portal he is building. He told the magazine he’s hoping “people will be able to get their polygenic scores for about as much as the cost of a cholesterol test.”
Some Experts Not Impressed with Broad Institute Study
But not all experts believe the Broad Institute/MGH/Harvard Medical School study deserves so much attention. Ali Torkamani, PhD, Director of Genomics and Genome Informatics at the Scripps Research Translational Institute, offered a tepid assessment of the Nature Genetics study.
In an article in GEN that noted polygenic risk scores were receiving “the type of attention reserved for groundbreaking science,” Torkamani said the recent news is “not particularly” a big leap forward in the field of polygenic risk prediction. He described the results as “not a methodological advance or even an unexpected result,” noting his own group had generated similar data for type 2 diabetes in their analysis of the UK dataset.
Nevertheless, Kathiresan is hopeful the study will advance disease treatment and prevention. “Ultimately, this is a new type of genetic risk factor,” he said in the news release. “We envision polygenic risk scores as a way to identify people at high or low risk for a disease, perhaps as early as birth, and then use that information to target interventions—either lifestyle modifications or treatments—to prevent disease.”
This latest research indicates healthcare providers could soon be incorporating polygenic risking scoring into routine clinical care. Not only would doing so mean another step forward in the advancement of precision medicine, but clinical laboratories and pathology groups also would have new tools to help diagnose disease and guide treatment decisions.
—Andrea Downing Peck
Related Information:
Genome-wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations
Predicting Risk for Common Deadly Diseases from Millions of Genetic Variants
Multigene Test May Find Risk for Heart Disease and More
A Harvard Scientist Thinks He Has a Gene Test for Heart Attack Risk. He Wants to Give It Away Free
Why Do Polygenic Risk Scores Get So Much Hype?
Oct 1, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
New advancements in mHealth, though encroaching on testing traditionally performed at clinical laboratories, offer opportunity to expand testing to remote locations
Mobile technology continues to impact clinical laboratories and anatomic pathology groups and is a major driver in precision medicine, as Dark Daily has reported. Most of the mobile-test development which incorporates smartphones as the testing device, however, has been for chemistry and immunoassay types of lab tests. Now, a new developer in Monmouth Junction, NJ, has created a Complete Blood Count (CBC) test that runs on devices attached to smartphones.
Such devices enable doctors to order test panels for patients in remote locations that also may lack resources, such as electricity.
The developer is Essenlix and it calls its new testing device iMOST (instant Mobile Self-Testing). According to the company’s website, which is mostly “Under Construction,” iMOST can provide “accurate blood and other healthcare testing in less than 60 seconds by a smartphone and matchbox-size-attachment, anywhere, anytime, and affordable to everyone.”
The company description on the Longitude Prize website states that Essenlix “uses multidisciplinary approaches to develop a new innovative platform of simple, fast, ultrasensitive, bio/chemical sensing and imaging for life science, diagnostics, and personal health.
The Longitude Prize competition was established to promote the invention of “an affordable, accurate, fast and easy-to-use test for bacterial infections that will allow health professionals worldwide to administer the right antibiotics at the right time,” the website states.

The Essenlix iMOST mobile-testing device (above) connects to a smartphone (shown right) and enables clinical laboratory technicians to run tests in remote locations from samples taken at time the test. Though still in trials, iMOST, and other similar devices, promise to expand testing to outside of traditional medical laboratory locations and further promote precision medicine. (Photos copyright: Lydia Ramsey/Business Insider.)
Essenlix’s iMOST mobile testing system consists of:
- a mobile application (app);
- the device attachment, which goes over the phone’s camera; and,
- a cartridge that holds a sample of blood.
So far, there have been two trials with a total of 92 participants, comparing traditional CBC testing with the Essenlix test. The results were within the FDA’s requirements for allowable error, prompting Chou to tell Business Insider, “Our error is clearly smaller than the FDA’s requirement, so the data is very, very good.”
Chou and his team are working toward FDA approval.
Other Testing Devices That Attached to Smartphones
Aydogan Ozcan, PhD, Professor of Electrical Engineering and Bioengineering at UCLA, and Mats Nilsson, PhD, Professor and Scientific Director of the Science for Life Laboratory at Stockholm University, have developed an attachment that they say can transform “a phone into a biomolecular analysis and diagnostics microscope,” according to The Pathologist. Dark Daily has published many e-briefings on Ozcan’s innovations over the years.
Their goal, the researchers said, was to create technology that can be used in low- and middle-income areas (LMICs), as well as in more advanced locations, such as Sweden. “I’ve been involved in other projects where we’ve looked at point-of-care diagnostic approaches,” he said, “and it seems to be very important that the devices [do not] rely on wired electricity or networks to serve not only LMICs, but also modern, developed environments. It’s often difficult to find an available power socket in Swedish hospitals.”
The molecular diagnostic tests that can be done with smartphone attachments—such as those developed by Ozcan and Nilsson—represent another way of using a smartphone in the healthcare arena, The Pathologist points out. Their invention combines the smartphone’s native camera, an app, optomechanical lasers, and an algorithm contained within the attachment to carry out fluorescence microscopy in the field.
Future of Mobile-Testing
An article appearing in the Financial Times describes some of the ways mobile technology is changing healthcare, including diagnostics that have traditionally been performed in the medical pathology laboratories.
“Doctors scan your body to look for irregularities, but they rely on pathologists in the lab to accurately diagnose any infection,” the article notes. “There, body fluids such as blood, urine, or spit are tested for lurking microbes or unexpected metabolites or chemicals wreaking havoc in your body. Now companies are miniaturizing these tests to create mobile pathology labs.”
Apple introduced the first iPhone in 2007. It’s doubtful anyone imagined the innovations in diagnostics and pathology that would soon follow. Thus, trying to predict what may be coming in coming decades—or even next year—would be futile. However, scientists and researchers themselves are indicating the direction development is headed.
Should Essenlix and other mobile-lab-test developers succeed in their efforts, it would represent yet another tectonic shift for medical pathology laboratories. Clinical laboratory managers and stakeholders should be ready, for the words of the ancient Greek philosopher Heraclitus have never been truer: “Change is the only constant in life.”
—Dava Stewart
Related Information:
Mobile Phone Microscopy
How Smartphones Are Transforming Healthcare
This Startup Wants to Make Blood Testing as Easy as Snapping a Photo with an iPhone
Is mHealth an Opportunity or Threat to Medical Laboratories and Pathology Groups?
New FDA Regulations of Clinical Decision-Support/Digital Health Applications and Medical Software Has Consequences for Medical Laboratories
UCLA Device Enables Diagnosis of Antimicrobial Resistance in Any Setting; Could Save Lives Lost to Antimicrobial Resistant Bacteria
Lab-on-a-Chip Diagnostics: When Will Clinical Laboratories See the Revolution?
Tiny, Simple-to-Use Lensless Microscope Might Soon Find a Place in Pathology
Sep 26, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
New metalens technology from MGH and SEAS researchers gives greater endoscopic optical imaging resolution and sample detail for anatomic pathologists performing diagnostics
Anatomic pathologists and clinical laboratories know that biopsy samples are necessary to diagnose many diseases. But, current endoscopic imaging techniques used by physicians sometimes fail to clearly visualize disease sites. Consequently, biopsies collected during these procedures may make it harder for pathologists and physicians to diagnose certain diseases and health conditions.
Now, a combined team of endoscopic imaging experts at Massachusetts General Hospital (MGH) and flat metalens developers at Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS) have developed “a new class of endoscopic imaging catheters—termed nano-optic endoscopes—that overcomes the limitations of current systems.”
That’s according to an article in Nature Photonics that reported on the research team’s study, published in Phys.org.
These nano-optics involved “flat metalenses” that promise to sharpen clarity and increase resolution of endoscopic imaging technology In turn, this contributes to more accurate pathology diagnostics and improve patient outcomes, while furthering the aims of precision medicine.
“Metalenses based on flat optics are a game changing new technology because the control of image distortions necessary for high resolution imaging is straightforward compared to conventional optics, which requires multiple complex shaped lenses,” Federico Capasso, PhD, Robert L. Wallace Professor of Applied Physics and Vinton Hayes Senior Research Fellow in Electrical Engineering at SEAS, and co-senior author of the study paper, told Nature Photonics. “I am confident that this will lead to a new class of optical systems and instruments with a broad range of applications in many areas of science and technology.”
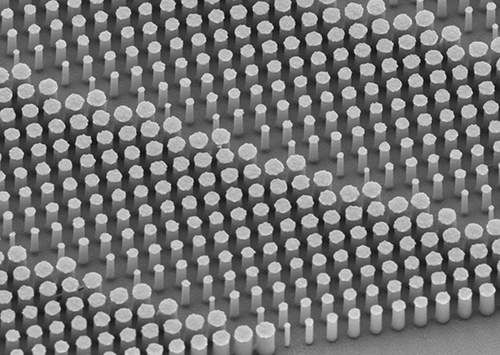
The image above shows a flat metalens taken using a scanning electron microscope. Anatomic pathologists and medical laboratories will benefit from the better quality biopsy specimens collected because of the sharper clarity and increased resolution of endoscopes built with the new nano-optics. (Photo copyright: Harvard SEAS.)
Researchers demonstrated the nano-optic endoscope’s ability to deeply penetrate and capture images at high resolutions in various tissues, including:
- Swine and sheep airways;
- Human lung tissue; and,
- Fruit flesh.
In the human lung tissue, “[T]he researchers were able to clearly identify structures that correspond to fine, irregular glands indicating the presence of adenocarcinoma, the most prominent type of lung cancer,” according to Phys.org.
Improving Endoscopic Imaging through Metalenses
The improved image resolution is due to the flat metalens configuration. “Currently, we are at the mercy of materials that we have no control over to design high-resolution lenses for imaging,” Yao-Wei Huang, PhD, Post-Doctoral Fellow at Harvard’s John A. Paulson School of Engineering and Applied Sciences and co-first author of the paper, told Phys.org.

Yao-Wei Huang, PhD (above), is a Post-Doctoral Fellow at Harvard’s John A. Paulson School of Engineering and Applied Sciences and co-first author of the study paper. “The main advantage of the metalens is that we can design and tailor its specifications to overcome spherical aberrations and astigmatism and achieve very fine focus of the light. As a result, we achieve very high resolution with extended depth of field without the need for complex optical components.” (Photo copyright: Harvard School of Engineering and Applied Sciences.)
The researchers note that current endoscopes using gradient-index (GRIN) lens-prism configurations and angle-polished ball lenses are used in a range of clinical applications due to their basic design. However, this benefit comes with shortfalls. “The ability of the nano-optic endoscope to obtain high-resolution images of sub-surface tissue structures in vivo is likely to increase the clinical utility of OCT [optical coherence tomography] in detection, diagnosis, and monitoring of diseases,” they state in their paper.
“The ability to control other properties of output light, such as the polarization state, enables a host of other applications—implausible to achieve using conventional catheters,” they continue. “Several tissues—such as smooth muscle, collagen (either innate or in fibrosis), and blood vessels—have constituent structures highly organized in one particular direction. Polarization-sensitive imaging can differentiate these structures from surrounding tissue by detecting their innate birefringence and optic axis.”
They further note that nano-optic endoscopes may yield benefits to other endoscopic optical imaging modalities such as confocal endomicroscopy.
Additional clinically-oriented studies will be required to assess how nano-optic endoscopes can elevate the capabilities of endoscopic OCT in examining fine pathological changes in luminal tissues.
Implications for Clinical Laboratories and Pathology Groups
The technology is still in the research stage with more trials needed to confirm the viability and accuracy of the approach. “This preclinical evaluation of the nano-optic endoscope indicated no significant flaws in the design for in vivo endoscopic imaging,” researchers note.
However, should nano-optic catheters gain clearance and change the endoscopy landscape as researchers predict, medical laboratories and pathologists might enjoy higher resolution images with greater information of the sample site—both key components of accurate diagnosis.
“Clinical adoption of many cutting-edge endoscopic microscopy modalities has been hampered due to the difficulty of designing miniature catheters that achieve the same image quality as bulky desktop microscopes,” Melissa Suter, PhD, Assistant Professor of Medicine at MGH and Harvard Medical School (HMS) and co-senior author of the study told Nature Photonics. “The use of nano-optic catheters that incorporate metalenses into their design will likely change the landscape of optical catheter design, resulting in a dramatic increase in the quality, resolution, and functionality of endoscopic microscopy. This will ultimately increase clinical utility by enabling more sophisticated assessment of cell and tissue microstructure in living patients.”
This research project at Massachusetts General Hospital and Harvard John A. Paulson School of Engineering and Applied Sciences is another example of how advances in technologies unrelated to surgical pathology can eventually contribute to improvements in how pathologists diagnose disease and help physicians identify the most promising therapies for their patients.
—Jon Stone
Related Information:
Nano-Optic Endoscope Sees Deep into Tissue at High Resolution
Nano-Optic Endoscope for High-Resolution Optical Coherence Tomography In Vivo
Nano-Optic Endoscope Allows High-Resolution Imaging
High-Resolution Nano-Optic Endoscope for Better Disease Detection