Aug 20, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
“On-a-chip” devices continue to advance and medical laboratories will be natural repositories for patient data as the technology continues to improve
Dark Daily has predicted that the future of clinical laboratory testing will include highly complex multi-analyte test panels. The biomarkers, however, could number in the hundreds or thousands. So, it’s interesting to see new research by a Massachusetts Institute of Technology (MIT) team currently developing a multi-biomarker organ test device for clinical purposes.
Motivated by the costly failure of animal testing efforts to develop drug safety and efficacy in humans, the MIT research engineers created a microfluidic platform technology they dubbed “physiome-on-a-chip,” or more colloquially, “body-on-a-chip.” Their goal is to identify drug reaction in different cell groups within the body (in vivo).
They acknowledged contributions of in vitro microphysiological systems (MPSs), AKA “organ-on-a-chip” (OOC) systems. They note, however, in their paper published in Scientific Reports, that more complex systems that interconnect and receive data from multiple MPSs are needed due to increasing limitations arising from drugs’ “lack of efficacy” rather than toxicity.
“Here we describe the development and implementation of multi-MPS platforms, AKA physiome-on-a-chip, supporting four-way, seven-way, and 10-way MPS interactions for several weeks,” the MIT engineers wrote.
Though MIT’s new technology needs further research and development time, as well as clinical trials, this type of chip design and its ability to scale is a positive development and progress toward Dark Daily’s prediction. Once finalized, it could be adopted in medical laboratories for many types of diagnostic testing purposes.
Researchers Motivated to Improve Drug Efficacy
According to an MIT news release, “MIT engineers have developed new technology that could be used to evaluate new drugs and detect possible side effects before the drugs are tested in humans. Using a microfluidic platform that connects engineered tissues from up to 10 organs, the researchers can accurately replicate human organ interactions for weeks at a time, allowing them to measure the effects of drugs on different parts of the body.”
The “body-on-a-chip” technology, MIT says, is aimed at determining how drugs may affect one organ while also having side effects on others.
“Some of these effects are really hard to predict from animal models because the situations that lead to them are idiosyncratic. With our chip, you can distribute a drug and then look for the effects on other tissues and measure the exposure and how it is metabolized,” said Linda Griffith, PhD, Professor of Teaching Innovation at MIT’s School of Engineering, and a senior author of the study, in the news release.
According to MIT, factors affecting the effectiveness of pharmaceuticals may include:
- Genetics;
- Environment;
- Personal lifestyles; and,
- Interactions with other drugs.
TechCrunch called the study “unprecedented,” pointing to the platform’s connection of so many tissues and the technology’s ability to keep them stable for weeks.

“An advantage of our platform is that we can scale it up or down and accommodate a lot of different configurations,” Linda Griffith, PhD, MIT Professor, MIT School of Engineering, told Science Daily. “I think the field is going to go through a transition where we start to get more information out of a three-organ or four-organ system, and it will start to become cost-competitive because the information you’re getting is so much more valuable.” (Photo copyright: MacArthur Foundation.)
How “Body-on-a-Chip” Works
“Body-on-a-chip” is about the size of a tablet computer and links 10 organ types, including: liver, lung, gut, endometrium, brain, heart, pancreas, kidney, skin, and skeletal muscle.
Using microfluidic platform technology, the researchers placed one- to two-million cells from human tissue samples into the device and then pushed fluid through the chip to resemble blood flow, the Daily Mail reported, adding that MIT’s MPS platform design features:
- Compartments made from a plastic block;
- Passages for fluid to move (as a circulatory system does) between the compartments;
- A water reservoir to limit fluid evaporation; and,
- Ability to monitor flow of molecular exchanges and drug distribution.
Essentially, using the MIT device, a drug can be introduced to one organ, processed normally, and then passed to other organs for processing and use in other ways, TechCrunch summarized.
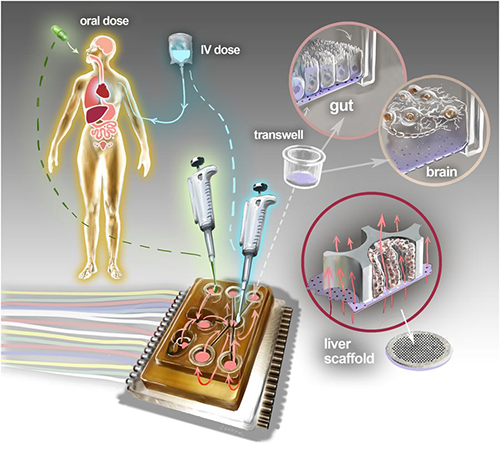
The physiome-on-a-chip system (above schematic) comprises bioengineered devices that nurture many interconnected 3D MPSs representing specified functional behaviors of each organ of interest, designed to capture essential features of in vivo physiology based on quantitative systems models tailored for individual applications such as drug fate or disease modeling. This technology could eventually be utilized for clinical laboratory and anatomic pathology testing. (Image and caption copyright: Victor O. Leshyk/Scientific Reports.)
Drug Delivery, Effects on Multiple Tissues Noted in MIT Study
The MIT researcher engineers reported these findings and accomplishments:
- Delivering a drug to the gastrointestinal tissue;
- Replicating digesting a drug;
- Observing as a drug was transported to other tissues and metabolized;
- Measuring a drug’s path; and,
- Noting effects of a drug on different tissues and how drugs break down.
“The huge potential of MPS technology is revealed by connecting multiple organ chips in an integrated system for in vitro pharmacology. This study beautifully illustrates that multi-MPS ‘physiome-on-a-chip’ approaches, which combine the genetic background of human cells with physiologically relevant tissue-to-media volumes, allow accurate prediction of drug pharmacokinetics and drug absorption, distribution, metabolism, and excretion,” said Kevin Healy, PhD, Professor of Bioengineering and Materials Science and Engineering, at University of California Berkeley in the MIT news release. Healy was not involved in the research.
Unique Device Design
In addition to making it possible to study so many different tissue types, the device design, according to MIT, is unique for these reasons:
- Its open microfluidic system, rather than a closed system, means the lid can be removed to manipulate tissue samples;
- Instead of external pumps common in closed systems, the MIT team used “on-board pumps” to control flow of liquid between the organs; and,
- The pumps used enabled larger engineered tissues, such as those from tumors in an organ, to be assessed.
The MIT engineers next plan to focus on specific organs—including the brain, liver, and gastrointestinal tissue—to model Parkinson’s disease, Digital Trends reported.
As healthcare providers and medical laboratories adopt precision medicine, MIT’s contributions are both timely and important. The ability to accommodate many different configurations in one platform is impressive, and something Dark Daily has been anticipating.
—Donna Marie Pocius
Related Information:
A “Body-on-a-Chip” Strings Together 10 Model Human Organs
“Body-on-a-Chip” Could Improve Drug Evaluation
MIT Builds “Body-on-a-Chip” Device That Can Store up to 10 Artificial Organs at Once
Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies
MIT Gadget Puts Multiple Artificial Organs into a Paperback-Sized Connected System
Drug Testing Could Get a Boost from MIT’s “Body-on-a-Chip”
Aug 15, 2018 | Digital Pathology, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Using precision genomics, Mayo researchers hope to develop improved medical laboratory tools for screening, diagnosing, and treating patients with inherited genetic disorders such as accelerated aging
Telomeres increasingly are on the radars of physicians and healthcare consumers alike, as researchers gain more knowledge about these critical nucleotides, and doctors continue to indicate their belief that telomeres could make useful diagnostic tools. If so, that would open up a new channel of precision medicine testing for clinical laboratories and anatomic pathology groups.
Telomeres are DNA strands that protect chromosome end points from degrading as people age. Their job is similar to the way plastic tips keep shoelaces from fraying, researchers at the Mayo Clinic explained in a news release. They have been using precision genomics in their assessment of 17 patients with short telomere syndrome (STS) to uncover the genetic causes of the condition.
They published their findings in the July issue of Mayo Clinic Proceedings.
Using Genetic Sequencing to Find Causes of Short Telomeres
People with STS could develop conditions including bone marrow failure, liver disease, and lung disease earlier in life than others, the news release pointed out.
However, according to the researchers’ paper, “Management of STSs is fraught with significant challenges such as delayed diagnoses, lack of routinely available diagnostics modalities, and standardized treatment guidelines.”
Nevertheless, some physicians are already leveraging information about telomeres in patient treatment. And many consumers have been turning to telomere diagnostic testing companies to learn the lengths of their own telomeres. They’ve learned that the longer the telomeres the better, as shorter telomeres are associated with accelerated aging.

“The length of certain telomeres gives a history of all the assaults a person has been subject to over the course of her lifetime,” a Wired article noted, quoting Joseph Raffaele, MD, co-founder of PhysioAge Medical Group, a clinical practice in New York City that specializes in “proactive” medicines. He goes on to call telomeres “the new cholesterol.” (Photo copyright: drraffaele.com.)
More Study into STS is Needed
GenomeWeb summarized the Mayo study’s methodology as follows:
Researchers reported these findings in Mayo Clinic Proceedings:
Study authors concluded that while some genetic mutations are common to short telomeres, they were found in only about 40% of the people in their study. So, more research is needed to discover other causes of short telomeres.
Telomeres and Lung Disease
Other research into telomeres was conducted by St. Paul’s Hospital and the University of British Columbia Department of Medicine, which focused on telomeres and lung disease.
In this study, researchers used polymerase chain reaction (PCR) to measure absolute telomere length from blood samples provided by 576 people with chronic obstructive pulmonary disease (COPD), according to a paper in the journal CHEST, published by the American College of Chest Physicians.
The study found that when compared to people with normal blood telomeres, people with shorter telomere lengths and more rapidly aging blood cells:
- Were 50% more likely to develop new or increasing respiratory symptoms;
- Were nine times more likely to die; and,
- Had worse health status and quality of life.
“It is known that short telomeres are associated with common morbidities of COPD, but it was not known if there is a relationship between blood telomeres and patient-related outcomes in COPD,” Don Sin, MD, a chest physician who led the research at the Centre for Heart Lung Innovation at St. Paul’s Hospital, stated in a news release.
Other Takes on Telomeres
A Harvard Medical blog noted, however, that short telomeres do not necessarily mean disease is imminent, nor that long ones guarantee optimal health.
“There is mounting evidence that a healthy lifestyle buffers your telomeres,” stated Immaculata De Vivo, PhD, a Harvard Medical School Professor and Genetics Researcher at the Dana-Farber/Harvard Cancer Center, in the blog post.
However, another expert questions the value of measuring telomeres for disease risk.
“In short, telomere lengths are too variable within a population, too variable within an individual, and too sensitive to environmental factors to offer any reliable information for common disease risk,” wrote Ricki Lewis, PhD, in PLOS.
Although there are many pitfalls to overcome, some doctors are pushing to use telomere information in patient treatment, and these studies from the Mayo Clinic and other researchers have contributed important data for diagnostic test developers.
In the end, vast and varied content about telomeres exists and clinical laboratory professionals may be called on to help clarify and assess the information. And that’s the long and the short of it.
—Donna Marie Pocius
Related Information:
Precision Genomics Point the Way to Mutations Associated with Accelerated Aging
Telomeres Are the New Cholesterol. Now What?
Clinical Correlates and Treatment Outcomes for Patients with Short Telomeres Syndrome
Mayo Clinic Researchers Use Targeted Sequencing to Diagnose Short Telomere Syndrome
Relationship of Absolute Telomere Length with Quality of Life, Exacerbations, and Mortality in COPD
Blood Telomeres Can Help Predict Risk of Disease Worsening or Death in COPD Patients
Can DNA Markers Called Telomeres Predict Aging?
Telomere Testing: Science or Snake Oil?
White Paper Download | How Next-Generation Sequencing Helps Molecular Laboratories Deliver Personalized Medicine Services to their Client Physicians
Summit: Breakthroughs with Genetic and Precision Medicine: What All Health Network CEOs Need to Know
Aug 13, 2018 | Laboratory Management and Operations, Laboratory News, Laboratory Pathology, Laboratory Testing
Without the beneficial bacteria, infants can develop gut dysbiosis, which can lead to severe chronic diseases
Another key insight into how the human microbiome performs essential functions has been discovered by a research team at the University of California, Davis (UCD). They have learned that nearly all babies born in developed nations no longer have a specific strain of bacteria called B. infantis, which digests a certain type of sugar found in breast milk.
Microbiologists, clinical laboratory scientist, and pathologists will find the UCD researcher’s discovery to be a fascinating insight into a newly-understood function of the human microbiome. Assuming that further research confirms these early findings, it also could lead to a medical laboratory assay for use during pregnancy or after delivery that would enable physicians to determine if the newborn is missing this strain of bacteria and what therapies would be appropriate.
Babies in Developed Nations Lack Beneficial B. infantis Bacteria
“The central benefits of having a microbiota dominated by B. infantis is that it crowds all the other guys out—especially pathogenic bacteria, which can cause both acute illnesses and chronic inflammation that leads to disease,” UC Davis researcher Bruce German, PhD, Professor and Chemist, Food Science and Technology, told the New York Times.
The UC Davis researchers published their study findings in mSphere, a journal of the American Society for Microbiology. In their paper they note that Bifidobacterium Infantis or B. infantis, a beneficial bacteria that aids in digestion, is missing from the microbiomes of infants in developed nations, such as the United States.
The study hypothesized that the reduction and eventual absence of B. infantis in American babies was the consequence of three factors:
- An increase in cesarean births;
- Use of commercial formulas instead of breast milk; and,
- Heightened use of antibiotics.
According to the New York Times, “Dr. German and his colleagues learned about the missing bacterium by studying breast milk. They found that the milk contains an abundance of oligosaccharides, carbohydrates that babies are incapable of digesting. Why would they be there if babies can’t digest them? They realized that these carbohydrates weren’t feeding the baby—they were feeding B. infantis.”
Good versus Bad Gut Bacteria
Because 70-80% of our immune system resides within our gastrointestinal tract, gut bacteria play an important role in our overall health. Breast milk contains essential probiotics and anti-inflammatory compounds that help “friendly” bacteria flourish in the infant gut.
There are more than two hundred different sugars or carbohydrates found in breast milk, known as human milk oligosaccharides (HMOs). They are one of the most copious components in breast milk but are completely indigestible by humans. So, why are they there?
Because they serve a critical role as food for microbes or prebiotics. Scientists have discovered that HMOs present in breast milk are there to feed the B. infantis, not to nourish the baby.
HMOs also act as a decoy to confuse undesirable bacteria from doing damage in the gut.
“Bad” bacteria are inclined to latch onto sugar molecules in intestinal cells. Because HMOs are very similar to those sugar molecules, the undesirable bacteria will instead latch onto the HMOs in a baby’s gut and leave vulnerable intestinal cells alone.
The primary benefits of B. infantis include:
- Production of short-chain fatty acids. When infantis digests HMOs, some short-chain fatty acids are released, which provide energy and help control yeast and fungus growth.
- Support for gut integrity. infantis signals gut cells in infants to generate proteins that fill gaps between intestinal cells. These gaps can be dangerous as they may allow toxins and bad bacteria to get into the bloodstream.
- Keeping undesirable bacteria at bay. infantis consumes HMOs and usurps space in the gut so potentially dangerous bacteria cannot take up residence or cause problems.
- Release of sialic acid. As it devours HMOs, infantis churns and releases sialic acid, a crucial nutrient for the brain development of infants.
- Production of folate. infantis also produces folate, which is necessary for infant development and growth and the creation of red blood cells.

“The need for clinicians to have a quick and reliable method to determine Bifidobacterium levels in [a] baby’s gut, and an effective way to replace the right Bifidobacterium to correct dysbiosis when detected, are the critical next steps for infant health,” noted Jennifer Smilowitz, PhD (above), Associate Director of Human Studies Research Program for the Foods for Health Institute at UC Davis, and one of the study authors, in a news release. (Photo copyright: UC Davis.)
The UC Davis study is the latest example of new insights about the microbiome, which refers to the collected genetic material of human microbiota. This promising field of research is expected to lead to a better understanding of how human gut bacteria affects resistance to certain chronic diseases, such as cancer, and to new clinical laboratory treatments and drug therapies.
Different research initiatives involving the human microbiome continue to indicate that gut bacteria can be a source of useful biomarkers for improving the health of individuals. Dark Daily has covered the study of human microbiome and development of new cancer therapies based on that research for many years.
Microbiome research, however, sometimes uncovers negative findings as well.
Lack of B. infantis, a principle gut microbe, can contribute to gut dysbiosis, which has been linked to chronic health conditions such as:
Researchers observed that reduction in B. infantis in the infant gut also has resulted in a rise in the pH of infant fecal matter. An analysis of 14 clinical studies performed between 1926 and 2017 showed a startling increase of pH from 5.0 to 6.5 in infant stools.
“These alarming changes to the infant gut microbiome and thus, gut environment, may be due to modern medical practices like antibiotics, C-sections, and formula feeding,” Jennifer Smilowitz, PhD, Associate Director of Human Studies Research Program for the Foods for Health Institute at UC Davis, and one of the study authors, noted in a news release. “These are all potentially life-saving medical practices but have unintended consequences on the infant gut microbiome. As a result, certain pathogenic bacteria—those linked to higher risk of health issues, such as colic, eczema, allergies, diabetes, and obesity—thrive.”
The process by which the researchers in this study identified the missing bacteria illustrates how more refined ways to examine molecules in the body are providing streamlined tools to identify elements within the body and their interaction with each other.
This new insight is one more confirmation that the human microbiome will be the source of useful diagnostic biomarkers, associated with medical laboratory therapies that can improve the health of individual patients.
—JP Schlingman
Related Information:
Elevated Fecal pH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century
The Bacteria Babies Need
Bifidobacterium Longum Subspecies Infantis: Champion Colonizer of the Infant Gut
Evolve BioSystems’ Activated B. infantis EVC001 Demonstrates Substantial and Persistent Remodeling of the Infant Gut Microbiome
How Baby’s First Microbes Could be Crucial to Future Health
Breast Milk and B. Infantis: Nature’s Favorite Probiotic
New Study Shows Significant Changes to Infant Fecal pH Over Last 100 Years
Researchers Discover Link between Gut Bacteria and the Effectiveness of Certain Cancer Drugs; Knowledge May Lead to New Types of Clinical Laboratory Tests
Researchers in Two Separate Studies Discover Gut Microbiome Can Affect Efficacy of Certain Cancer Drugs; Will Findings Lead to a New Clinical Laboratory Test?
Microbiologists at Weill Cornell Use Next-Generation Gene Sequencing to Map the Microbiome of New York City Subways
Aug 10, 2018 | Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
This low-cost, reusable noninvasive light test could serve as a prototype for detecting other biomarkers and diseases in rural and outlying medical laboratories
A 24-year-old Ugandan computer scientist whose own malaria was missed by traditional clinical laboratory blood tests has developed a device that detects signs of the disease using a beam of light directed onto a patient’s finger. The light highlights and detects changes in the color, shape, and concentration of red bloods cells affected by disease.
Brian Gitta, CEO and co-founder of computer software company thinkIT Limited, became the youngest winner of the UK’s Royal Academy of Engineering Africa Prize for Engineering Innovation. His eHealth solution is called Matibabu, which means “treatment” in Swahili.
Gitta and his team are developing a low-cost, reusable device that clips onto a patient’s finger and provides malaria test results within 60 seconds through a mobile phone app, UPI.com reported. The latest Matibabu prototype uses a ‘hybrid of magnetic-optic technology and electro-impedance technology’ to detect the disease,” according to a blog post on the thinkIT website.
“Our next step is to determine the validity and reliability of the Matibabu device compared with the gold standard microscopy and RDT by conducting field tests with malaria patients in selected health facilities in order to obtain information that will be used to improve the device, and eventually roll it out to the market,” the blog post states.

The Matibabu malaria detection device (above) uses the principles of light scattering and magnetism to detect changes to red blood cells that signal disease. The low-cost, reusable device from thinkIT Limited has advanced through several prototypes and now has an estimated 80% accuracy rate. (Photo copyright: Makerere University College of Engineering, Design, Art and Technology.)
TechCrunch reported that the current generation of Matibabu is about 80% accurate, with the expectation that further development will increase the device’s accuracy to 90-95%.
While this new diagnostic technology needs further development and clinical studies, it could potentially be used for other biomarkers and diseases besides malaria. However, according to the Centers for Disease Control and Prevention (CDC), rapid diagnostic tests (RDTs) like this are not yet widely used, so speed-of-diagnosis also is an issue.
Nevertheless, if successful, such a non-invasive test for malaria would be a major breakthrough since, today, the mosquito-borne disease must be confirmed by medical laboratory blood tests using either microscopic diagnosis or antigen detection, which are costly and time consuming.
“It’s a perfect example of how engineering can unlock development—in this case by improving healthcare,” Rebecca Enonchong, Africa Prize judge, noted in a Royal Academy of Engineering statement. “Matibabu is simply a gamechanger.”

Africa Prize judge Rebecca Enonchong (left) presents Ugandan Brian Gitta (right) of Matibabu with the Africa Prize winner’s medal. (Photo/caption copyright: Royal Academy of Engineering.)
Shafik Sekitto, Matibabu Vice President of Business Development, told BBC World News that Gitta’s own battle with malaria was prolonged because the first three blood tests failed to diagnose his disease. “[Gitta] brought up the idea: ‘Why can’t we find a new way of using the skills we have in computer science—of diagnosing a disease without having to prick somebody?’” Sekitto explained.
Malaria Threatens Half the World’s Population
The World Health Organization (WHO) estimates that nearly half the world’s population is at risk of malaria. According to WHO estimates, in 2015 there were 429,000 deaths worldwide from malaria, with 90% of cases and 92% of deaths in sub-Saharan Africa.
The Africa Prize, which includes a $33,400 (124-million Uganda shillings) award, is Africa’s biggest prize dedicated to engineering innovation. Its sponsors aim to encourage engineers from sub-Saharan Africa “to apply their skills to develop scalable solutions to local challenges.” In addition to funding, award recipients also receive business training, mentoring, and access to the Royal Academy of Engineering’s network of high profile and experienced engineers and experts, and their networks.
Gitta expects the award of the Africa Prize will help thinkIT Limited better navigate the difficult process of gaining worldwide regulatory approval for a new diagnostic device.
“It’s such a big achievement for us, because it means that we can better manage production in order to scale clinical trials and prove ourselves to regulators,” he predicted in the Royal Academy of Engineering statement. “The recognition will help us open up partnership opportunities—which is what we need most at the moment.”
Many pathologists and clinical laboratory managers are watching the efforts of various companies to develop medical laboratory tests that can be performed with a device that is coupled to a smartphone and can be performed as a point-of-care test. A substantial proportion of these research efforts are targeting the needs for accurate diagnostic testing in developing countries. That’s because of the need for cheap, fast, and accurate assays that can be performed in the rural areas of these nations.
—Andrea Downing Peck
Related Information:
Ugandan Inventor Wins Africa Prize for Bloodless Malaria Test
Ugandan Innovation Wins the Africa Prize for Engineering Innovation
Ugandan Wins Africa Prize for Bloodless Malaria Test
Matibabu Uses Light to Diagnose Malaria
Matibabu Wins the Africa Prize for Engineering Innovation
Aug 8, 2018 | Compliance, Legal, and Malpractice, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing, Management & Operations
As federal regulatory agencies continue to push transparency, hospitals, medical labs, anatomic pathology groups, and other healthcare providers must develop strategies for remaining competitive and maintaining patient volume
More price transparency continues to be a goal of Medicare officials. Some clinical laboratory managers and pathologists may be unaware of a proposed rule issued by the federal Centers for Medicare and Medicaid Services (CMS) in April that would require hospitals to post prices online as early as this January 1, 2019.
This action is a definitive demonstration of how the federal healthcare program continues to scrutinize how healthcare organizations communicate with patients and post their prices. This pressure on healthcare systems and individual providers extends to the clinical laboratories and anatomic pathology groups that support them. Without a plan to address these changes, medical laboratory test volume and practice revenues can be severely impacted.
Recent coverage from RevCycleIntelligence indicates that this trend is only just getting started.
“When you go to receive a healthcare service, there are always going to be situations where you can’t know what the costs will be, especially around emergency situations and some acute situations,” Centers for Medicare and Medicaid (CMS) Administrator Seema Verma told RevCycleIntelligence. “But for a lot of us, we’re going in for planned procedures. You should be able to know what it’s going to cost you.”
In April 2018, CMS posted a proposed rule that requires hospitals to post standard service rates online and update their pricing lists at least annually starting January 1, 2019. While many healthcare organizations currently report pricing to state boards and other online directories, this rule, if enacted, could make it easier for consumers to source reliable pricing information before obtaining care.
“We are concerned that challenges continue to exist for patients due to insufficient price transparency. Such challenges include patients being surprised by out-of-network bills for physicians—such as anesthesiologists and radiologists who provide services at in-network hospitals—and patients being surprised by facility fees and physician fees for emergency room visits. We also are concerned that chargemaster data are not helpful to patients for determining what they are likely to pay for a particular service or hospital stay,” the CMS proposed rule states.
In the proposed rule that references out-of-network bills, pathologists should have been included as a hospital-based physician service—such as anesthesiologists and radiologists—that submits bills to patients for care provided in the hospital. Pathology practice administrators may want to review the specific language of the proposed rule to understand how hospitals served by the pathology group will be required to post prices.

“If you’re buying a car or pretty much anything else, you’re able to do some research,” Seema Verma (above), Administrator, Centers for Medicare and Medicaid Services told RevCycleIntelligence. “You’re able to know what the quality is. You’re able to make comparisons. Why shouldn’t we be able to do that in healthcare? Every healthcare consumer wants that.” (Photo copyright: Centers for Medicare and Medicaid Services.)
Transparency Concerns Lead to Additional Questions from CMS
Alongside the proposed rule, CMS also is issuing a request for information (RFI) to give healthcare experts an opportunity to:
- Demonstrate the impact of current proposals; and,
- Make recommendations to further align the proposal with both the needs of healthcare organizations and service providers as well as the cost-reduction goals of CMS.
Key questions, according to RevCycleIntelligence, include:
- How should “standard charges” be defined (e.g., average or median rates for chargemaster items; average or median rates for groups of services commonly billed together as determined by the hospital; or average discount off the chargemaster amount across all payers)?
- What types of information would help patients understand hospital prices and patient financial responsibility? How should hospitals use this information to inform patients and decision-making?
- Should healthcare providers be required to tell patients about their out-of-pocket costs for a service prior to care delivery? Should providers even play a role in informing patients of out-of-pocket costs?
- Should CMS require providers to give patients information on what Medicare pays for a service?
- How should CMS enforce healthcare price transparency requirements? Should hospitals have to attest to meeting requirements?
Implications for Healthcare Services and Diagnostics Providers
RevCycleIntelligence reports that between CMS rules and bills introduced by Senators, an increase in healthcare transparency pricing is likely. These requirements will continue to apply pressure to clinical laboratories, anatomic pathology groups, and other diagnostics providers.
Much like the importance of communicating value to the new wave of payer and physician partnerships emerging around the country, transparency will offer an opportunity to communicate value to consumers.
However—particularly for high-margin services and assays—laboratories must create strategies to address pricing transparency and communicate the value of their services if they hope to maintain volumes and financial integrity at existing levels.
A study conducted by Johns Hopkins University researchers and published in The American Surgeon also highlights benefits that transparency might hold outside of simple regulatory compliance.
Analyzing data from six ambulatory care centers from 2016, they found that five out of six reported increases in both patient volume and revenue after adopting price transparency. Half of the centers also reported a reduction in administrative burden—a concern that medical laboratories must also address as streamlining operations and optimizing efficiency becomes a core part of successful lab operation in the face of healthcare reform.
Clinical laboratories and anatomic pathology groups should develop a strategy for addressing new transparency requirements. That strategy should include ways to effectively communicate their value to both healthcare providers and consumers. Should the CMS proposed rule progress to a final rule, failure to address pricing transparency may result in enforcement and compliance concerns—a critical issue for laboratories already facing tighter markets and increased regulatory and payer scrutiny.
—Jon Stone
Related Information:
“Just the Beginning” of Healthcare Price Transparency, Verma Says
CMS Aims to Catalyze Advancements in Consumer Price Transparency
Verma: CMS Will “Use Every Lever” for Promoting Interoperability, Data Access
CMS to Require Healthcare Price Transparency Online for Hospitals
Publish Your Prices, Boost Your Bottom Line
Healthcare Price Transparency in U.S. Not Improved in Recent Years
The Impact of Price Transparency for Surgical Services
Patients’ Views on Price Shopping and Price Transparency
Aug 6, 2018 | Digital Pathology, Instruments & Equipment, Laboratory Instruments & Laboratory Equipment, Laboratory Management and Operations, Laboratory News, Laboratory Operations, Laboratory Pathology, Laboratory Testing
Researchers in Boston are working to develop DNA as a low-cost, effective way to store data; could lead to new molecular technology industries outside of healthcare
Even as new insights about the role of DNA in various human diseases and health conditions continue to tumble out of research labs, a potential new use for DNA is emerging. A research team in Boston is exploring how to use DNA as a low-cost, reliable way to store and retrieve data.
This has implications for the nation’s clinical laboratories and anatomic pathology groups, because they are gaining experience in sequencing DNA, then storing that data for analysis and use in clinical care settings. If a way to use DNA as a data storage methodology was to become reality, it can be expected that medical laboratories will have the skillsets, experience, and information technology infrastructure already in place to offer a DNA-based data storage service. This would be particularly true for patient data and healthcare data.
Finding a way to reduce the cost of data storage is a primary reason why scientists are looking at ways that DNA could be used as a data storage technology. These scientists and technology developers seek ways to alleviate the world’s over-crowded hard drives, cloud servers, and databases. They hope this can be done by developing technologies that store digital information in artificially-made versions of DNA molecules.
The research so far suggests DNA data storage could be used to store data more effectively than existing data storage solutions. If this proves true, DNA-based data storage technologies could play a key role in industries outside of healthcare.
If so, practical knowledge of DNA handling and storage would be critical to these companies’ success. In turn, this could present unique opportunities for medical laboratory professionals.
DNA Data Storage: Durable but Costly
Besides enormous capacity, DNA-based data storage technology offers durability and long shelf life in a compact footprint, compared to other data storage mediums.
“DNA has an information-storage density several orders of magnitude higher than any other known storage technology,” Victor Zhirnov, PhD, Chief Scientist and Director, Semiconductor Research Corporation, told Wired.
However, projected costs are quite high, due to the cost of writing the information into the DNA. However, Catalog Technologies Inc. of Boston thinks it has a solution.
Rather than producing billions of unique bits of DNA, as Microsoft did while developing its own DNA data storage solution, Catalog’s approach is to “cheaply generate large quantities of just a few different DNA molecules, none longer than 30 base pairs. Then [use] billions of enzymatic reactions to encode information into the recombination patterns of those prefab bits of DNA. Instead of mapping one bit to one base pair, bits are arranged in multidimensional matrices, and sets of molecules represent their locations in each matrix.”
The Boston-based company plans to launch an industrial-scale DNA data storage service using a machine that can daily write a terabyte of data by leveraging 500-trillion DNA molecules, according to Wired. Potential customers include the entertainment industry, federal government, and information technology developers.
Catalog is supported by $9 million from investors. However, it is not the only company working on this. Microsoft and other companies are reportedly working on DNA storage projects as well.

“It’s a new generation of information storage technology that’s got a million times the information density, compared to flash storage. You can shrink down entire data centers into shoeboxes of DNA,” Catalog’s CEO, Hyunjun Park, PhD (above center, between Chief Science Officer Devin Leake on left and Milena Lazova, scientist, on right), told the Boston Globe. (Photo copyright: Catalog.)
Microsoft, University of Washington’s Synthetic DNA Data Storage
Microsoft and researchers at the University of Washington (UW) made progress on their development of a DNA-based storage system for digital data, according to a news release. What makes their work unique, they say, is the large-scale storage of synthetic DNA (200 megabytes) along with the ability to the retrieve data as needed.
“Synthetic DNA is durable and can encode digital data with high density, making it an attractive medium for data storage. However, recovering stored data on a large-scale currently requires all the DNA in a pool to be sequenced, even if only a subset of the information needs to be extracted,” the researchers wrote in their paper published in Nature Biotechnology.
“Here, we encode and store 35 distinct files (over 200 megabytes of data ) in more than 13-million DNA oligonucleotides and show that we can recover each file individually and with no errors, using a random access approach,” the researchers explained.
“Our work reduces the effort, both in sequencing capacity and in processing, to completely recover information stored in DNA,” Sergey Yekhanin, PhD, Microsoft Senior Researcher, told Digital Trends.
Successful research by Catalog, Microsoft, and others may soon lead to the launch of marketable DNA data storage services. And medical laboratory professionals who already know the code—the life code that is—will likely find themselves more marketable as well!
—Donna Marie Pocius
Related Information:
The Rise of DNA Data Storage
The Next Big Thing in Data Storage is Actually Microscopic
Catalog Hauls in $9 Million to Make DNA-Based Data Storage Commercially Viable
UW and Microsoft Researchers Achieve Random Access in Large-Scale DNA Data Storage
Random Access in Large-Scale DNA Data Storage
Microsoft and University of Washington Show DNA Can Store Data in Practical Way











